
Computational methods and opportunities for phosphorylation network medicine
Introduction
Phosphorylation, the addition of the phosphate group, PO43– to a protein substrate by protein kinases, is one of the most common post-translational modifications (PTM) of proteins. Phosphorylation plays a central role in cell signaling and regulatory mechanisms, and can lead to activation or inhibition of downstream signaling events, depending on the substrates and involved pathways. Based on the nature of the phosphorylated –OH group, these kinases are classified as protein-serine, threonine or tyrosine kinases. The dysregulation of these kinases, or the resulting change in phosphorylation events, is a potential signaling mechanism involved in cancer development and progression. One such example is P53, a well-known tumor suppressor, which has been observed to have a wide range of PTM, including multi-site phosphorylation, acetylation, methylation and ubiquitylation (1) suggesting extensive control of the activity of this protein. Another tightly regulated example is SRC (and SRC family kinase proteins). These proto-oncogenic proteins are known to be essential for cell differentiation, motility, proliferation, and survival (2). In addition to the complexity of multi-site phosphorylation by protein kinases, autophosphorylation creates internal regulation of activity and increases the overall complexity of the regulation process.
In addition to the essential role that phosphorylation plays in signaling, regulation, and physiology, another reason that research on phosphorylation has become significant in cancer research is because kinases have become one of the largest classes of drug targets (3). Major progress has been made against some types of cancers using inhibitors of tyrosine kinases as therapeutic agents. Often these kinases are altered or activated in cancer, making inhibition of their activity specific to cancer cells and thus targetable. The use of imatinib in chronic myeloid leukemia (CML) patients with the presence of the BCR-Abl tyrosine kinase is one of the earlier examples of targeted therapeutics and often cited as a paradigm for research in cancer therapeutics (4,5). Many additional kinase inhibitors have been developed. Several well-known examples are: gefitinib/erlotinib for epidermal growth factor receptor (EGFR) mutant driven lung cancers, and crizotinib for EML4-ALK driven lung cancers, and vemurafenib for melanoma patients with BRAF V600E mutation. Furthermore, rather than targeting a single protein or gene, there is an increasing recognition of the importance of potential therapeutic strategies on targeting interacting molecules, modular domains, or a biological network as a whole (6).
The advancement of proteomics technologies within cancer research enables the quantitation of phosphorylation. In this review, section I briefly outlines how (phosphorylated) peptides are identified and quantified, and what computational issues and challenges remain. In section II, we describe several phosphoproteomics databases and online resources for annotation of detected phosphorylation events. In section III, we describe and compare several popular computational tools and resources for phosphorylation network inference and reconstruction. This field is not new but has evolved quickly, as the evolution of the proteomics platforms generating the data have resulted in increasingly accurate, quantifiable proteomics data. Our discussion is focused on recent developments in analytical approaches. In section IV, we summarize some computational approaches linking phosphorylation or kinases to medicine. In the final section, we describe open questions and research opportunities for moving the field forward.
I. Quantitative phosphoproteomics
Mass spectrometry (MS) has been used for the identification of proteins through digested peptides and modifications of these peptides, such as phosphorylation, through database search techniques. Qualitative data (e.g., presence of a specific phosphosite) is an important step towards network inferences however this information limits possible approaches. Increasingly, it is becoming possible to quantify the abundance of peptides, proteins and phosphosites within complex mixtures in a high-dimensional way. The development of new experimental techniques and new software has enabled automated approaches to quantitative phosphoproteomics. We present here a brief overview of approaches taken for identification and quantification of phosphoproteins from LC-MS/MS experiments.
Phosphosite identification
When phosphoproteomics experiments using LC-MS/MS are undertaken, a common result is a list of identified peptides from trypsin-digested proteins in the sample. By including database searches for PTM’s (such as phosphorylation), the sequences of peptides can be inferred from peptide fragmentation. In the case of unmodified peptide sequences, this approach can be very accurately performed. However, in the case of modifications the peptides may be correctly identified but it may be difficult to identify the specific site that was observed to be phosphorylated. Depending on the fragmentation patterns and the number of possible sites, there may be multiple locations for a phosphorylation to have occurred. For instance, in the case of tyrosine phosphorylation, it is possible to have two (or more) tyrosines in the same peptide and thus determining which tyrosine is phosphorylated is not clear. More detailed discussion of this topic can be found in (7). Moreover, digestion of peptides is not always complete or there may be multiple available locations for cleavage. In this case, there may be several different peptides which contain the phosphosite in question. Thus in the case of quantitative phosphoproteomics, quantifying specific peptides must be further processed to accumulate measurements for the particular phosphosite. The sum or average of peptide quantities can be used to summarize measurements to a specific site.
Phosphoproteomics quantification methods
Quantification of proteins from MS experiments has been of interest for a number of studies. There are several approaches to quantification, including the use of isotope labeling of samples or conditions in order to identify relative quantifications [e.g., SILAC (8,9) or iTRAQ (10)]. It is possible that each technique provides some additional information for understanding proteomics profiles of samples (11). More challenging is the use of label-free quantification (LFQ), in which labeled proteins are not used but the peak area, centroid or other measure of abundance in the m/z MS1 spectra is considered.
Several types of quantification approaches exist for phosphoproteomics and are similar to those discussed in (12). The simplest approach is spectral counting (13) of peptides (or phosphosites) and is a frequently employed approach to generate abundance information. The number of spectra (or MS2 scans) in which the peptide has been identified is used as a surrogate of the abundance of that peptide (13). As described previously, the situation is complicated by the need to count spectra for peptides that are partially overlapping but also contain the phosphosite. Several numerical issues arise from this type of processing, including the exponential nature of peptide detection. Many peptides are seen once in a single experiment, whereas it is relatively infrequent for a peptide to be seen many times. This process of detection is inherently stochastic therefore the reliability of the measurements is debated. However, this approach is very simple and broadly used. A number of tools have been developed utilizing spectral counting to estimate protein abundance, including the crux spectral-counts command, implemented as part of the Crux software toolkit (14). A review of spectral counting is (15).
Over the last several years, a number of software packages have been developed that automate the process of extracting quantification of specific peptides from LC-MS/MS data. This type of quantification without isotope labeling is typically termed LFQ. For more discussion on the rationale behind LFQ, see for example (16). Peaks in the chromatographic profile are used to estimate abundance. This process can be very time-consuming manually but with software that can align profiles across samples with the MS2 identification, large numbers of proteins per experiment can be estimated. A number of tools have been developed, including LFQuant (17), MaxQuant (18), SuperHirn (19), and an open-MS based pipeline (20). MaxQuant software, developed as an integrated solution, incorporates many of the necessary steps for quantification in the same package. This software performs peak detection in MS1 spectra and aggregates information over retention times to estimate peak intensities. Peptide identification is then performed, currently using Andromeda (21). Peptides can be combined, particularly in the case of overlapping peptides that cover the same phosphosite. The MaxQuant software also supports isotope labeled experiments such as SILAC (stable amino acid isotope-label) data. A comparison of quantification approaches can be read in (22). Although less challenging in phosphoproteomics data, the task of combining individual peptide quantifications into protein measurements has been tackled by a number of investigators. These approaches are reviewed in (23). We have summarized the discussed quantification methods in Table 1.
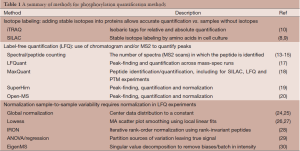
Full table
Normalization
Data generated by high-dimensional Omics technologies are often affected by a variety of known or unknown systematic biases (31). Proteomics technologies are no exception. In quantitative proteomics datasets, these biases or batch effects, i.e., non-biological signals, may occur due to variations in sample processing steps, different instrument performance in different days or batches, or differences of experimental conditions (32). Typically, the data is first logarithm (base two) transformed, and then followed by normalization to eliminate or avoid the systematic bias or batch effects. After log transformation, the distribution is approximately normally distributed, and the base two makes it easy for interpretation. For instance, a difference of 1 in log2 scale is a two-fold change in the original scale. Since phosphoproteomics are often quantified using MS-based technologies, we summarize several common normalization methods for LC-MS based data although these are not just specific to phosphoproteomics data.
A global normalization is one of the simplest approaches, and it often works well. The goal is to center the distribution of the log transformed values to a constant value for each sample. For instance, ‘quality control samples’ (QC samples) can be planned in between biological samples of interest within each batch of samples. Each batch is composed of a consecutive run of 10 or 20 samples followed by multiple preparations of the same QC samples. These QC samples are used to monitor potential systematic batch effects. When a global normalization method is used, it could be centered to the median of the same QC samples across batches in the entire project (24), or just a simple mean, median, or a fixed constant for each sample (25). A constant value could also be estimated using a subset of peptides from house-keeping proteins when such proteins are identified.
The need for normalization is not proteomics-specific, but often found in a wide variety of high-throughput platforms. Some of the normalization methods, such as those developed for RNA expression microarrays, are also used to normalize proteomics data. For instance, a popular scatter plot smoothing method known as lowess regression is a commonly applied normalization method developed for microarray platforms (26), and also applied to proteomics data (27). Scatterplot smoothing uses the “MA plots” for comparing the intensities of two samples. The techniques were originally developed for two-color cDNA expression microarrays, in which there is an internal reference sample to compare against. Lowess performs local linear fits on the user-defined fraction of points to be used for smoothing, and some optimization based methods for estimating the fraction have been proposed (33). With the assumptions that a portion of the genes are rank-invariant between samples, other normalization methods are possible. An example is iterative rank-order normalization (IRON), developed by our group for microarray normalization. IRON uses the best-performing techniques from each of several popular processing methods while retaining the ability to incrementally renormalize data without altering previously normalized expression (28). ANOVA and regression models are another popular way to partition out explicitly each source of variations in which that treatment terms and batches are commonly factors in the models (29). Modified from original methods for microarray normalization (34), EigenMS, was developed and uses singular value decomposition to capture and remove biases from LC-MS peak intensity measurements (30). This allows for bias to be captured as eigen peptides and then removed. The advantage is that if the batch effects are not always explicitly modeled, they could still be estimated and removed. It has been our experience that a one-size fits all solution is overly simplistic in terms of which normalization to use, due to different systematic biases introduced in different experiments. However, the need for normalization and the potential existence of batch effects is important to acknowledge. Given a reasonable study design and appropriate normalization, most batch effects can be eliminated.
II. Annotating phosphorylation sites
The use of phosphoproteomics in recent years has been enabled through an extensive development of the appropriate technologies (35). Increasingly, there is demand for computational inference of cellular regulatory mechanisms using these phosphoproteomics measurements. However, central to the ability for inferring relationships is the capability to catalog data from experimental conditions in which instances of the relationships have been observed. These data are generated from different organisms, such as yeast, bacteria and plants (36) or vertebrates, mammals or a variety of species. Databases have been developed for different organisms for depositing these annotated data. An excellent review of databases developed for this purpose for various organisms can be found in (37). We summarize updated information on three large comprehensive human- or vertebrate-centric phosphoproteomics databases in Table 2. Briefly, PhosphoSitePlus (PSP) is a database with a comprehensive collection of different PTMs including phosphorylation, ubiquitinylation, acetylation and methylation (41) created by cell signaling technology (CST). Among all included PTMs, 78% are phosphorylation, 15% ubiquitinylation, 6% acetylation. One feature PSP has is its ability to generate high-through phosphoproteomics data, including both low-throughput and high-throughput experimental data, and rapid sharing the newly generated data through their website. As of 2/27/2014, there are 210,352 phosphosites are available via their website (http://www.phosphosite.org/homeAction.do). In contrast, the Phospho.ELM resource (http://phospho.elm.eu.org/) focuses primarily on the collection of manually curated phosphosites and information derived from small-scale experiments (38-40). It stores information on ~300 kinases and over 8,000 substrates, over 42,000 phosphosites. The Human Protein Reference Database (HPRD) is one of the largest databases for the human proteome (42) (http://www.hprd.org/). It includes protein-protein interactions (PPI), PTMs, enzyme/substrate relationships, disease associations, tissue expression, and subcellular localization of human proteins. For non-commercial usage, the data is freely downloadable. It currently has information on 95,016 phosphosites mapped to 13,041 proteins. It has been emphasized that the information are all carefully and manually curated (instead of generated by automated data mining algorithms). Among the 95,016 annotated phosphosites, 88,250 of them are identified based on in vivo analysis alone, 2,678 from in vitro experiments and 4,088 phosphosites by both methods. Furthermore, only 5,930 phosphorylation events, i.e., kinase-substrate relationships (KSR) have been curated. Only a very small fraction of the total observed phosphorylation events were curated. In other words, for the vast majority of the identified in vivo phosphosites, the specific kinase(s) responsible for the phosphorylation events remain unknown. Computational approaches to infer KSRs introduced below attempt to bridge this wide gap.
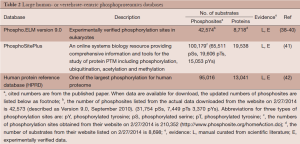
Full table
III. Phosphorylation network inference
Many different computational approaches have been taken for reconstructing phosphorylation networks, or connecting kinases and phosphosites into a biological signaling network. The first is a common approach to annotate phosphoproteins to pathways, modules, or protein-protein interaction networks using existing knowledge in pathway or network databases, such as Ingenuity Pathway Analysis (www.ingenuity.com), PANTHER (43,44), KEGG (45), GeneGo metaCore pathway analysis (www.genego.com), or STRING (46). There is no dynamic information in this kind of annotation, but a snap shot of all possible interactions.
The second approach, although not mutually exclusive from the first approach, has been primarily focused on inferring the phosphorylation of substrates by corresponding kinases, commonly referred as KSR. Development of computational methods to predict binding substrate specificities of protein kinases started from experimental identification of consensus sequence motifs recognized by the active sites of kinases (47,48). However, these sequence motifs often lack sufficient information to uniquely identify substrates of specific kinases. For example, the sites phosphorylated by different kinases from the CDK and SRC families cannot be distinguished by their consensus sequences alone. Several of these general computational approaches can be applied to enhance the evidence and narrow down to key players in the signaling network. For instance, Kim et al. has dissected the signaling network of TBK1, an emerging drug target using phosphoproteomics data (49) by applying multiple analyses, including gene ontology (GO) pathway enrichment analysis, motif analysis using the motif-x algorithm (50), and GeneGo MetaCore pathway analysis. Table 3 provides an overview of the statistical methods or bioinformatics tools described in this section for inferring phosphorylation network.
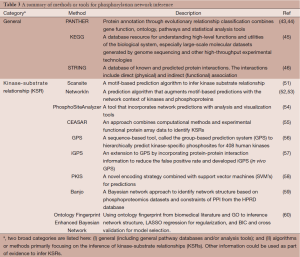
Full table
Network context of kinases and phosphoproteins are incorporated by some of prediction algorithms such as NetworKIN (52,53). The contextual information includes subcellular compartmentalization, co-localization via anchoring proteins and scaffolds, and temporal and cell-type specific co-expression. The algorithm uses neural networks and position-specific scoring matrices to assign phosphosites to one or more kinase families, based on the intrinsic preference of kinases for consensus substrate motifs (51) at the first stage. In the second stage, the contextual information from the STRING database is incorporated to improve the specificity from the motif-based predictions. It has reconstructed the phosphorylation network with 7,143 site-specific interactions and improves the prediction accuracy of known KSRs from 25% to 64% (52). The data is available for download. In their follow-up paper focusing on their online database, it is indicated that there are 20,224 site-specific interactions available through their website involving 3,978 phosphoproteins and 73 human kinases from 20 families (53).
Building on predictions made from NetworKIN, downstream analysis can be performed. While NetworKIN is a powerful tool for predictions both the interface (web-based tool) and the licensing limit the flexibility to perform such analysis. PhosphoSiteAnalyzer (54) is one of the first tools to incorporate NetworKIN predictions with analysis tools. Kinase predictions are extracted from NetworKIN using automated software to “screen scrape” results from the NetworKIN website using C# scripts. The process is still semi-automated since user interaction is required to save result files locally. Input files for PhosphoSiteAnalyzer include FASTA files for protein sequences, specific phosphosite positions (used for kinase predictions), and additional annotation about the experimental conditions (e.g., responsive to a compound, log ratios, P values from a statistical test). Once the predictions are generated and extracted from NetworKIN, a variety of analyses are available that typically involve characterizing the kinases that are predicted to be phosphorylating proteins. For instance, there are subset-specific kinase enrichment analyses which combine experimental annotation (e.g., class labels) with the predicted kinases for enrichment among a subgroup (as determined by a Fisher exact test). Consensus motifs in kinase-phosphorylation interactions can be compared between subgroups, suggesting a specific motif. Network analysis of KSR can be performed in Cytoscape (61) after exporting the connections from the software. Additionally, kinase-kinase relationships can be identified using clustering and heat maps of co-occurrence. PhosphoSiteAnalyzer provides an integrated environment for motif-enrichment analysis and kinase associations based on quantitative data.
Different proteomics platforms have been developed and applied to study phosphorylation. A recently developed approach, CEASAR (55), attempts to combine computational approaches and experimental data to identify KSRs using functional protein arrays. The authors first identified the proteins on the array that could be phosphorylated by a given kinase and then removed the false positive signals using control experiments. The collection of 289 kinases and 1,967 substrates after step 1 is referred to as the “rawKSR” dataset. The second step to enrich for physiological relevance is performed by integrating the contextual information by applying a Bayesian model (62). Using a positive training set composed of 1,103 experimentally validated kinase-substrate pairs; and a negative training set with 10,000 proteins containing no known kinases, a likelihood L score for each of the 24,046 KSRs was calculated. Applying a P value of 0.05 as threshold, a refined 3,656 KSRs involving 255 unique proteins and 742 substrates were predicted and referred to as “refKSRs”. The third step combines additional known and curated KSRs and generates the combined dataset, referred to as comKSR. The forth step uses an iterative motif prediction approach to combine the rawKSR and in vivo phosphosites. After the final step, a phosphorylation map with 4,417 KSRs, connecting between 2,591 sites from 652 substrates and 230 kinases was generated, and is referred to as a high-resolution map of human phosphorylation networks. Among the 7,143 site-specific KSRs identified by NetworKIN, after removing the kinases or substrates with obsolete Ensembl IDs, 6,336 site-specific kinase substrate interactions remain. Newman et al. (55) used the known 1,156 site-specific KSRs as the bench mark to evaluate the performance of CEASAR against NetworKIN. The true positive rate of NetworKIN is 0.76% (48/6,336) while CEASAR has 17.2% true positive rate (758/4,417) with a >20-fold improvement in the true positive rate. This might not be surprising since the known 1,156 site-specific KSRs, which were used to compare the performance, likely include all 1,103 interactions used as part of the positive training dataset in step 3. The authors primarily credited the improvement to using full length proteins, instead of using peptides. Fully folded protein structures could potentially be important for substrate recognition.
Xue et al. have developed a sequence-based tool, called the group-based prediction system (GPS) to hierarchically predict kinase-specific phosphosites for 408 human kinases (56). Xue and his collaborators later extended their algorithms to incorporate protein-protein interaction information to reduce the false positive rate and developed iGPS (in vivo GPS) (57). They constructed eukaryotic phosphorylation networks and predicted a total of 186,922 site-specific KSRs from 1,079 proteins kinases and 9,247 substrates for 44,290 phosphosites in five different species, including yeast, C. elegans, drosophila, mouse and human. The stand-alone applications are freely available for download (http://gps.biocuckoo.org/). Currently, on their website, it is indicated that GPS 3.0 can predict KSRs for 464 human kinases.
Another approach, PKIS (58), uses a novel encoding strategy combined with support vector machines (SVMs) to predict a KSR. As reported by these authors, Phospho.ELM currently lists 3,151 KSRs. However, this is less than the 12% of the total 27,404 sites in the database thus arguing the need for additional work. The predictor for PKIS was trained to predict specific kinases interacting with specific phosphosites. It is trained on a subset of kinases from Phospho.ELM that had at least ten predicted phospho sites. Similar to GPS-related methods, there is the inclusion of negative examples. That is, examples are provided in which kinases phosphorylate sites which are phosphorylated by other kinases (not the target kinase). This is distinguished from the more common approach of examples in which a site is selected that is not phosphorylated at all. This approach allows the classifier to see both positive examples of kinase-phosphorylation relationships and negative examples of no relationship between kinase and phosphorylation. The sequence in the kinase was encoded using an occurrence frequency of each amino acid within a 30AA window. The authors demonstrate successful prediction of a set of kinase-phosphorylation relationships identified using CEASER (55). The results of predictions were generally high in specificity and similar in sensitivity as other methods, including GPS2.1, Musite and others.
Bayesian network analysis provides a probabilistic graph-based approach to model signaling pathways (63), account for the uncertainties (64), and to reconstruct signaling networks (65,66). A Bayesian network is a directed acyclic graph (DAG), in which the nodes are variables and the edges are dependence relations between two given nodes. For instance, Banjo (59) was used to search and identify network structure based on phosphoproteomics datasets. Due to a vast possible number of network structures, the following biological constraints were used to constrain the search (66): any interactions must be PPI from HPRD, the proteins must be connected to at least one protein in the HPRD database, the proteins have to be measured at least in two out of the four datasets, which the authors have measured in their datasets. Other approaches using Bayesian networks also exist. For instance, Ontology Fingerprint Enhanced Bayesian Network (60) first used the ontology fingerprint from biomedical literature and GO, which is a set of GO terms overrepresented in the PubMed abstracts linked to a gene (or phenotype) and its enrichment statistical significance, evaluated by P values (67). Based on the network structure, the search process then used the information on pair-wise similarity of ontology fingerprints to decide to add or remove edges, and to generate large numbers of candidate networks. The authors trained the networks based on the proteomics data using the expectation-maximization algorithm with LASSO regression for regularization. Finally, BIC and cross-validation were used to select the best predicted network in the training dataset. The results showed that their method seemed to be able to capture signal transduction and predicted phosphorylation levels with high correlation (R2 =0.93). Partial least-squares regression (PLSR) was used to reduce the dimensions and also evaluate the optimal number of partial least squares components to retain using root-mean-squared error (RMSE) (68).
There are also more statistical or mathematical models developed for inferring signaling networks or regulatory networks using transcriptomics data. This is partly due to the fact that microarray gene expression data are widely available. Although describing methods for inferring networks of other types (other than phosphorylation network) is not within the scope of this review, however, some of the methods could be modified and applied to infer phosphorylation networks. Network component analysis decomposes a matrix into two matrices, one for the connectivity strength and the other for the regulatory signals. It uncovers hidden regulatory signals from outputs of a network when partial knowledge is available (69). We also have developed a Bayesian method by incorporating information on pathways and gene networks in the analysis of DNA microarray data to select markers, which are associated with survival outcome in a breast cancer study (70). Prior pathway information was used to define pathway summaries, specify prior distributions, and structure the Markov chain Monte Carlo (MCMC) moves to fit the model. In a separate work on inferring the regulatory relationship between miRNA and mRNAs, instead of selecting predictive markers, i.e., the nodes in a network, we used a Bayesian graphical model to select the regulatory relationships, i.e., the edges, between miRNA and mRNAs (71). We will discuss more open questions including some of the extensions of this work in the final section.
IV. Connecting kinases or phosphorylation to medicine
Kinases have become one of the largest ‘druggable’ groups in cancer therapeutics in recent years. Various computational approaches were applied or developed to connect kinases or phosphorylation events to medicine, for potential therapeutic strategies or biomarker identification for drug response. For instance, unsupervised cluster analyses were applied to the phosphotyrosine profiling of tyrosine kinases from 41 non-small cell lung cancer (NSCLC) cell lines and ~150 NSCLC tumors. The authors have identified existing known oncogenic kinases such as EGFR and c-Met as well as (at the time) novel drivers, such as ALK and ROS fusion proteins (72). For selecting phosphoproteomics markers for predicting Dasatinib response, Wilcoxon rank sum tests were first performed, followed by leave-one-out cross-validation to decide the number of markers/features to keep in the model, and a SVM with linear kernel was used for building the final classifier (73). Protein-protein interaction networks were visualized after the model building. However, the network information was not used for selection of the signature of 12 phosphosites in this study. A recent tool, HyperModules, was developed identifying clinically and phenotypically associated network modules with disease mutations for biomarker discovery (74). The kinase-substrate network was used in the algorithm.
Given a set of kinases with mutations, people are interested in finding out what potential kinase inhibitors could be used for cancer treatment. Recently, large scale kinase inhibitor selectivity profiles have become available (75,76). K-MAP (77), an online tool using these two kinase inhibition profiles (75,76) as reference databases, was developed to correlate and prioritize kinase inhibitors that enriches a set of query kinases provided by users. In the first reference database, Anastassiadis et al. systematically investigated 178 commercially available inhibitors against a panel of 300 protein kinases and assessed the kinase inhibition (IC50) (75) while Davis et al. evaluated the inhibitor selectivity and potency of 72 inhibitors on 442 kinases using direct binding affinities between kinases and inhibitors (Kd) (76). The pattern matching approach is similar to the Connectivity Map (78), which uses Kolmogorov-Smirnov (KS) statistics. The “score” reported on the website is normalized between 0 and 1 for each drug and inhibitors are ranked based on this normalized score. A permutation-based P value is based on randomly permuting the total number of kinases in each query from the ranked order list for each drug. A user-friendly web-based tool, implemented using Python, is available (http://tanlab.ucdenver.edu/kMap).
V. Open opportunities
Phosphorylation at different phosphosites of a protein could lead to different molecular events and physiological responses. For instance, phosphorylation at different tyrosine sites of the SRC kinase could lead to either activation or inhibition of the SRC kinase (79). Different kinase inhibitor sensitivities were found among different sites of EGFR (17). Identifying and quantification of phosphorylation events associated with drug response to the site-specific level remains non-trivial and will be informative for characterizing the drug mechanism and their potential clinical utilities.
Important functional roles for phosphorylation and other PTMs have been known for decades, but only in the past 10 years has MS-based proteomics begun to reveal the extent of the PTM universe (80). The rapid evolution and development of new proteomics technologies pose challenges and also provide great research opportunities on how to quickly develop computational tools and statistical methods to analyze and incorporate data generated from these new platforms. An example of an emerging platform is the activity based protein profiling (ABPP) technique, which enables the quantification of a proteome-level landscape of kinase activities (81,82). A number of enzyme families have been studied using this approach, including serine hydrolases, kinases, phosphatases, and metalloproteinases. ATP-based probes have been used to tag and enable LC-MS/MS based identification and quantification of protein kinases and other important ATP-binding proteins in cancers cells. A large degree of the kinome can be captured (~80%) and effects of kinase inhibitors can be profiled. This kind of experimental data could be integrated with existing network knowledge in the public database or literature.
There is an increasing recognition that understanding network interaction and interplay is essential to develop effective therapeutic strategies for treating complex diseases, such as cancer (6,83). As discussed in section III, phosphorylation network reconstruction has been a main research interest in the computational community, and improvement on phosphorylation network inference has been made in the past decade. Similarly, there has been proof of concept work on drug repurposing by connecting kinases to therapeutics using computational approaches (77,84) (section IV). Figure 1 shows a schematic representation of how a generalized linear regression model could be set up to integrate the phosphorylation network information and utilize the quantitative phosphoproteomics data to select targets and drugs computationally (Figure 1). The recent work of HyperModules is an example (74) of utilizing phosphorylation information for biomarker discovery. Other models could also be developed. For instance, Bayesian models incorporating pathways and network information could be modified for incorporating a phosphorylation network and/or protein-protein interaction network. We have previously developed an approach to select pathways and genes simultaneously on gene expression data through stochastic search to predict breast cancer survival outcome (70) and could be modified for this purpose. Furthermore, combination treatment strategies have become more common in recent years. There are attempts to select combinations of treatments using network information (85), however there remains an open question as to what an effective informatics or statistical method is to identify effective combination therapeutics. Biology is complex; the interplay between de-phosphorylation by phosphatases and phosphorylation by kinases, as well as the role of auto-phosphorylation, is not discussed in this review but clearly adds complexity to this type of analysis.
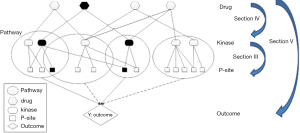
There are many applications for the developments within phosphorylation network medicine. It is known that chemical compounds or drugs can interact with multiple and/or unintended targets including kinases. Drug-repurposing or repositioning has been one of the active areas in the past several years, suggesting that some medicines can be used to treat other (unintended) diseases. In addition, drug-drug interaction (DDI) is an emerging threat to public health. Recent estimates indicate that DDIs cause nearly 74,000 emergency room visits and 195,000 hospitalizations each year in the USA (86). The adverse effects due to side effect of drugs or DDI are clearly an area for significant contribution by developments described in this review. Most gene-drug relationships are contained within the scientific literature, but are dispersed over a large number of publications, with thousands of new publications added each month. There has been successful computational work that serves as a proof of principle on automated text mining as a potential solution for identifying gene-drug relationships and aggregating them to predict novel DDIs (87). Research on using computational approaches to extract DDI or side effects with an emphasis on kinase inhibitor and phosphoproteomics knowledge for cancer therapeutics is largely under explored. Thus we believe that computational developments in understanding and constructing the signaling networks from phosphoproteomics will play a significant role in human health within cancer and beyond.
Acknowledgments
Special thanks to the phosphoproteomics research group at the Moffitt Cancer Center: Eric Haura, John Koomen, Uwe Rix, and Keiran Smalley. We also thank Alvaro Monteiro, Kate Fisher, Eric Welsh, Jiannong Li, and Guolin Zhang for the helpful discussion.
Funding: The work is funded by the Anna D. Valentine USF-Moffitt Cancer Research Award.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Translational Cancer Research for the series “Statistical and Bioinformatics Applications in Biomedical Omics Research”. The article has undergone external peer review.
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.3978/j.issn.2218-676X.2014.05.07). The series “Statistical and Bioinformatics Applications in Biomedical Omics Research” was commissioned by the editorial office without any funding or sponsorship. YAC served as the unpaid Guest Editor of the series. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Loughery J, Meek D. Switching on p53: an essential role for protein phosphorylation. BioDiscovery 2013;8:1:20.http://www.biodiscoveryjournal.co.uk/Article/10.7750/BioDiscovery.2013.8.1#.U4dDevldWcK
- Roskoski R Jr. Src protein-tyrosine kinase structure and regulation. Biochem Biophys Res Commun 2004;324:1155-64. [PubMed]
- Cohen P. Protein kinases--the major drug targets of the twenty-first century? Nat Rev Drug Discov 2002;1:309-15. [PubMed]
- Druker BJ, David A. Karnofsky Award lecture. Imatinib as a paradigm of targeted therapies. J Clin Oncol 2003;21:239s-245s. [PubMed]
- Stegmeier F, Warmuth M, Sellers WR, et al. Targeted cancer therapies in the twenty-first century: lessons from imatinib. Clin Pharmacol Ther 2010;87:543-52. [PubMed]
- Haura EB. From modules to medicine: How modular domains and their associated networks can enable personalized medicine. FEBS Letters 2012;586:2580-5. [PubMed]
- Wiese H, Kuhlmann K, Wiese S, et al. Comparison of Alternative MS/MS and Bioinformatics Approaches for Confident Phosphorylation Site Localization. J Proteome Res 2014;13:1128-37. [PubMed]
- Ong SE, Blagoev B, Kratchmarova I, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics 2002;1:376-86. [PubMed]
- Ong SE, Kratchmarova I, Mann M. Properties of 13C-substituted arginine in stable isotope labeling by amino acids in cell culture (SILAC). J Proteome Res 2003;2:173-81. [PubMed]
- Ross PL, Huang YN, Marchese JN, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics 2004;3:1154-69. [PubMed]
- Merl J, Ueffing M, Hauck SM, et al. Direct comparison of MS-based label-free and SILAC quantitative proteome profiling strategies in primary retinal Muller cells. PROTEOMICS 2012;12:1902-11. [PubMed]
- Mueller LN, Brusniak MY, Mani DR, et al. An assessment of software solutions for the analysis of mass spectrometry based quantitative proteomics data. J Proteome Res 2008;7:51-61. [PubMed]
- Liu H, Sadygov RG, Yates JR 3rd. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem 2004;76:4193-201. [PubMed]
- McIlwain S, Mathews M, Bereman MS, et al. Estimating relative abundances of proteins from shotgun proteomics data. BMC Bioinformatics 2012;13:308. [PubMed]
- Lundgren DH, Hwang SI, Wu L, et al. Role of spectral counting in quantitative proteomics. Expert Rev Proteomics 2010;7:39-53. [PubMed]
- America AH, Cordewener JH. Comparative LC-MS: a landscape of peaks and valleys. PROTEOMICS 2008;8:731-49. [PubMed]
- Zhang W, Zhang J, Xu C, et al. LFQuant: a label-free fast quantitative analysis tool for high-resolution LC-MS/MS proteomics data. Proteomics 2012;12:3475-84. [PubMed]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 2008;26:1367-72. [PubMed]
- Mueller LN, Rinner O, Schmidt A, et al. SuperHirn - a novel tool for high resolution LC-MS-based peptide/protein profiling. Proteomics 2007;7:3470-80. [PubMed]
- Weisser H, Nahnsen S, Grossmann J, et al. An Automated Pipeline for High-Throughput Label-Free Quantitative Proteomics. J Proteome Res 2013;12:1628-44. [PubMed]
- Cox J, Neuhauser N, Michalski A, et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 2011;10:1794-805. [PubMed]
- Trudgian DC, Ridlova G, Fischer R, et al. Comparative evaluation of label-free SINQ normalized spectral index quantitation in the central proteomics facilities pipeline. Proteomics 2011;11:2790-7. [PubMed]
- Matzke MM, Brown JN, Gritsenko MA, et al. A comparative analysis of computational approaches to relative protein quantification using peptide peak intensities in label-free LC-MS proteomics experiments. Proteomics 2013;13:493-503. [PubMed]
- Shan L, Chen YA, Davis L, et al. Measurement of phospholipids may improve diagnostic accuracy in ovarian cancer. PloS One 2012;7:e46846 [PubMed]
- Callister SJ, Barry RC, Adkins JN, et al. Normalization approaches for removing systematic biases associated with mass spectrometry and label-free proteomics. J Proteome Res 2006;5:277-86. [PubMed]
- Bolstad BM, Irizarry RA, Astrand M, et al. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003;19:185-93. [PubMed]
- Ting L, Cowley MJ, Hoon SL, et al. Normalization and Statistical Analysis of Quantitative Proteomics Data Generated by Metabolic Labeling. Mol Cell Proteomics 2009;8:2227-42. [PubMed]
- Welsh EA, Eschrich SA, Berglund AE, et al. Iterative rank-order normalization of gene expression microarray data. BMC Bioinformatics 2013;14:153. [PubMed]
- Hill EG, Schwacke JH, Comte-Walters S, et al. A statistical model for iTRAQ data analysis. J Proteome Res 2008;7:3091-101. [PubMed]
- Karpievitch YV, Taverner T, Adkins JN, et al. Normalization of peak intensities in bottom-up MS-based proteomics using singular value decomposition. Bioinformatics 2009;25:2573-80. [PubMed]
- Leek JT, Scharpf RB, Bravo HC, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet 2010;11:733-9. [PubMed]
- Karpievitch YV, Dabney AR, Smith RD. Normalization and missing value imputation for label-free LC-MS analysis. BMC Bioinformatics 2012;13:S5. [PubMed]
- Berger JA, Hautaniemi S, Jarvinen AK, et al. Optimized LOWESS normalization parameter selection for DNA microarray data. BMC Bioinformatics 2004;5:194. [PubMed]
- Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet 2007;3:1724-35. [PubMed]
- Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature 2003;422:198-207. [PubMed]
- Gribskov M, Fana F, Harper J, et al. PlantsP: a functional genomics database for plant phosphorylation. Nucleic Acids Research 2001;29:111-3. [PubMed]
- Xue Y, Gao X, Cao J, et al. A summary of computational resources for protein phosphorylation. Curr Protein Pept Sci 2010;11:485-96. [PubMed]
- Diella F, Cameron S, Gemund C, et al. Phospho.ELM: A database of experimentally verified phosphorylation sites in eukaryotic proteins. BMC Bioinformatics 2004;5:79. [PubMed]
- Diella F, Gould CM, Chica C, et al. Phospho.ELM: a database of phosphorylation sites--update 2008. Nucleic Acids Res 2008;36:D240-4. [PubMed]
- Dinkel H, Chica C, Via A, et al. Phospho.ELM: a database of phosphorylation sites—update 2011. Nucleic Acids Research 2011;39:D261-7. [PubMed]
- Hornbeck PV, Kornhauser JM, Tkachev S, et al. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res 2012;40:D261-70. [PubMed]
- Goel R, Harsha HC, Pandey A, et al. Human Protein Reference Database and Human Proteinpedia as resources for phosphoproteome analysis. Mol Biosyst 2012;8:453-63. [PubMed]
- Mi H, Muruganujan A, Casagrande JT, et al. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 2013;8:1551-66. [PubMed]
- Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res 2013;41:D377-86. [PubMed]
- Kanehisa M, Goto S, Sato Y, et al. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 2012;40:D109-14. [PubMed]
- von Mering C, Jensen LJ, Snel B, et al. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res 2005;33:D433-7. [PubMed]
- Hjerrild M, Stensballe A, Rasmussen TE, et al. Identification of Phosphorylation Sites in Protein Kinase A Substrates Using Artificial Neural Networks and Mass Spectrometry. J Proteome Res 2004;3:426-33. [PubMed]
- Puntervoll P, Linding R, Gemünd C, et al. ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Research 2003;31:3625-30. [PubMed]
- Kim JY, Welsh EA, Oguz U, et al. Dissection of TBK1 signaling via phosphoproteomics in lung cancer cells. Proc Natl Acad Sci U S A 2013;110:12414-9. [PubMed]
- Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol 2005;23:1391-8. [PubMed]
- Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res 2003;31:3635-41. [PubMed]
- Linding R, Jensen LJ, Ostheimer GJ, et al. Systematic discovery of in vivo phosphorylation networks. Cell 2007;129:1415-26. [PubMed]
- Linding R, Jensen LJ, Pasculescu A, et al. NetworKIN: a resource for exploring cellular phosphorylation networks. Nucleic Acids Res 2008;36:D695-9. [PubMed]
- Bennetzen MV, Cox J, Mann M, et al. PhosphoSiteAnalyzer: a bioinformatic platform for deciphering phospho proteomes using kinase predictions retrieved from NetworKIN. J Proteome Res 2012;11:3480-6. [PubMed]
- Newman RH, Hu J, Rho HS, et al. Construction of human activity-based phosphorylation networks. Mol Syst Biol 2013;9:655. [PubMed]
- Xue Y, Ren J, Gao X, et al. GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol Cell Proteomics 2008;7:1598-608. [PubMed]
- Song C, Ye M, Liu Z, et al. Systematic analysis of protein phosphorylation networks from phosphoproteomic data. Mol Cell Proteomics 2012;11:1070-83. [PubMed]
- Zou L, Wang M, Shen Y, et al. PKIS: computational identification of protein kinases for experimentally discovered protein phosphorylation sites. BMC Bioinformatics 2013;14:247. [PubMed]
- Yu J, Smith VA, Wang PP, et al. Advances to Bayesian network inference for generating causal networks from observational biological data. Bioinformatics 2004;20:3594-603. [PubMed]
- Qin T, Tsoi LC, Sims KJ, et al. Signaling network prediction by the Ontology Fingerprint enhanced Bayesian network. BMC Syst Biol 2012;6:S3. [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [PubMed]
- Hu J, Wan J, Hackler L, et al. Computational analysis of tissue-specific gene networks: application to murine retinal functional studies. Bioinformatics 2010;26:2289-97. [PubMed]
- Needham CJ, Bradford JR, Bulpitt AJ, et al. Inference in Bayesian networks. Nat Biotechnol 2006;24:51-3. [PubMed]
- Pe’er D. Bayesian network analysis of signaling networks: a primer. Sci STKE 2005;2005:pl4.
- Woolf PJ, Prudhomme W, Daheron L, et al. Bayesian analysis of signaling networks governing embryonic stem cell fate decisions. Bioinformatics 2005;21:741-53. [PubMed]
- Bose R, Molina H, Patterson AS, et al. Phosphoproteomic analysis of Her2/neu signaling and inhibition. Proc Natl Acad Sci 2006;103:9773-8. [PubMed]
- Tsoi LC, Boehnke M, Klein RL, et al. Evaluation of genome-wide association study results through development of ontology fingerprints. Bioinformatics 2009;25:1314-20. [PubMed]
- Janes KA, Albeck JG, Gaudet S, et al. A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science 2005;310:1646-53. [PubMed]
- Liao JC, Boscolo R, Yang YL, et al. Network component analysis: Reconstruction of regulatory signals in biological systems. Proc Natl Acad Sci 2003;100:15522-7. [PubMed]
- Stingo FC, Chen YA, Tadesse MG, et al. Incorporating Biological Information into Linear Models: A Bayesian Approach to the Selection of Pathways and Genes. Ann Appl Stat 2011;5:1978-2002. [PubMed]
- Stingo FC, Chen YA, Vannucci M, et al. A Bayesian Graphical Modeling Approach to MicroRNA Regulatory Network Inference. Ann Appl Stat 2010;4:2024-48. [PubMed]
- Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 2007;131:1190-203. [PubMed]
- Klammer M, Kaminski M, Zedler A, et al. Phosphosignature Predicts Dasatinib Response in Non-small Cell Lung Cancer. Mol Cell Proteomics 2012;11:651-68. [PubMed]
- Leung A, Bader GD, Reimand J. HyperModules: identifying clinically and phenotypically significant network modules with disease mutations for biomarker discovery. Bioinformatics 2014; [PubMed]
- Anastassiadis T, Deacon SW, Devarajan K, et al. Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat Biotechnol 2011;29:1039-45. [PubMed]
- Davis MI, Hunt JP, Herrgard S, et al. Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol 2011;29:1046-51. [PubMed]
- Sullivan KD, Padilla-Just N, Henry RE, et al. ATM and MET kinases are synthetic lethal with nongenotoxic activation of p53. Nat Chem Biol 2012;8:646-54. [PubMed]
- Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 2006;313:1929-35. [PubMed]
- Roskoski R. Src kinase regulation by phosphorylation and dephosphorylation. Biochem Biophys Res Commun 2005;331:1-14. [PubMed]
- Olsen JV, Mann M. Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol Cell Proteomics 2013;12:3444-52. [PubMed]
- Patricelli MP, Nomanbhoy TK, Wu J, et al. In situ kinase profiling reveals functionally relevant properties of native kinases. Chem Biol 2011;18:699-710. [PubMed]
- Patricelli MP, Szardenings AK, Liyanage M, et al. Functional interrogation of the kinome using nucleotide acyl phosphates. Biochemistry 2007;46:350-8. [PubMed]
- Pawson T, Linding R. Network medicine. FEBS Letters 2008;582:1266-70. [PubMed]
- Kim J, Yoo M, Kang J, et al. K-Map: connecting kinases with therapeutics for drug repurposing and development. Hum Genomics 2013;7:20. [PubMed]
- Azmi AS, Wang Z, Philip PA, et al. Proof of concept: network and systems biology approaches aid in the discovery of potent anticancer drug combinations. Mol Cancer Ther 2010;9:3137-44. [PubMed]
- Percha B, Altman RB. Informatics confronts drug–drug interactions. Trends Pharmacol Sci 2013;34:178-84. [PubMed]
- Percha B, Garten Y, Altman RB. Discovery and explanation of drug-drug interactions via text mining. Pac Symp Biocomput 2012;410-21. [PubMed]


