Nanoformulation enhances anti-angiogenic efficacy of tunicamycin
Introduction
Gold nanoparticles (Au NPs) and peptide-based nanostructures have been receiving significant attention over the past decade due to their potential applications in catalysis, chemical sensing, electronics, optics, sensors and biomedical applications (1-4). Particularly, monolayer-protected Au NPs using thiolated compounds to stabilize Au NPs have been gaining popularity for delivering various therapeutic agents such as drugs, proteins, and nucleic acids into their targets (5-7). Several types of Au NP conjugates have now been prepared for potential drug delivery applications. For example, Corbierre et al. (8) has recently reported polystyrene-functionalized Au NPs by the covalent attachment of thiol-terminated polystyrene prepared by anionic polymerization. Synthesis of Au NPs with tetra (ethylene glycol) ylated cationic ligands, fluorogenic ligands as well as polycaprolactone-methoxy poly (ethylene glycol) has also been carried out for the development of drug delivery systems (9,10). In a separate study, NP-polymer transfection vectors have been synthesized as well (11,12). While Au NPs provide a versatile platform for the preparation of drug delivery devices, peptide based nanotubes self-assembled from peptide bolaamphiphiles (amino acid head groups covalently bound via hydrocarbon chain), exhibit several properties that make them promising biomaterial candidates, including facile self-assembly in aqueous solutions and adaptability to functionalization for increased biocompatibility (13-20). Furthermore, the peptide head groups can be readily modified in order to manipulate and potentially alter the properties of the self-assembled micro- and nanostructures. Although many applications related to peptide-based nanotubes have been investigated (21-25), the immense potential of peptide nanotubes as drug delivery devices is yet to be fully tapped.
The objective of this manuscript is to evaluate if nano-formulation of Tunicamycin would enhance its efficacy. The rational is that nano-structure particles will evade the immune system’s clearing mechanisms long enough to reach the targeted disease tissue efficiently. Tunicamycin (a Mr840 dalton glucosamine-containing pyrimidine nucleoside and a biologic), a competitive inhibitor of N-acetylglucosaminyl 1-phosphate transferase (GPT) has recently been shown to inhibit (I) angiogenesis in vitro and in vivo; (II) the breast tumor microvasculature; and (III) prevent the breast tumor progression in athymic nude mice (26). Tunicamycin was effective in treating indiscriminately the double- and triple-negative breast tumors. The effect is mediated by “ER stress” followed by developing unfolded protein response (upr) in tumor microvasculature and induction of apoptosis.
Breast cancer is a multi-factorial disease which depends not only on the genetic makeup but also on the metabolomic profile of the individual as well as on the environment. A great diversity in breast cancer incidence rate suggests both endogenous and exogenous factors contribute to the development and progression of the disease. The etiology of the disease is complex, but this hyperproliferative disorder is angiogenesis dependent (27-31). The “angiogenic switch” (32) is therefore a critical process in which the dynamic balance between pro- and anti-angiogenic factors is shifted to the former by conditions created by the tumor and its microenvironment, including hypoxia, inflammation, and mutation in oncogenes or tumor suppressor genes, such as p53. Commonly known pro-angiogenic factors are vascular endothelial growth factor (VEGF), basic fibroblast growth factor (FGF-2), epidermal growth factor (EGF), platelet-derived growth factor (PDGF), placental growth factor (PIGF), and matrix metalloproteinases (MMPs). Endogenous anti-angiogenic factors include thrombospondin (TSP-1), angiostatin, tumstatin, and endostatin (33,34).
The prognosis of breast cancer depends upon the stage at diagnosis. Each year, breast cancer is diagnosed in over one million women worldwide, with a survival rate of approximately 400,000 women (35). The breast cancer statistics for 2013 indicates the following in the United States: 232,400 new cases of invasive breast cancer, 64,640 new cases of carcinoma in situ (CIS; non-invasive), and nearly 39,620 deaths.
Normal vasculature is quiescent in healthy adults with each endothelial cell dividing once every 10 years. In contrast, tissue remodeling and angiogenesis are crucial for the growth and metastasis of breast cancer, providing an attractive therapeutic target (36). Treatment strategies therefore include either (I) targeting angiogenesis with endothelial toxins, growth factor antagonists, protease inhibitors, endogenous anti-angiogenics, anti-angiogenic chemotherapy; or (II) other targets.
Several lines of evidence now indicate that N-linked glycoproteins play an important role in capillary endothelial cell proliferation and differentiation (37-45). Deoxymannojirimycin (an inhibitor of hybrid and complex-type N-glycans), inhibited the formation of capillary tubes when tested in vitro by plating capillary endothelial cells on fibronectin-coated dishes. In contrast, swainsonine (an inhibitor of complex- but not hybrid-type N-glycans), did not inhibit tube formation.
In this study, we have tested various nano-formulations of Tunicamycin (e.g., nanoparticles, bound to Au NPs, encapsulated in peptide nanotubes containing threonine moieties, etc.) as an alternative drug delivery device for breast cancer glycotherapy. The reason being certain drug such as doxorubicin (DXR) has been found to be more effective when conjugated to hydrophilic nanoparticles that penetrate more deeply into the cell than the drugs alone (46). The nano-formulations of Tunicamycin are not only developed for the first time here but they have been studied for the first time as well. The expectation is that nano-formulated Tunicamycin will have the potential to enhance the efficacy of the drug, due to the high surface to volume ratio of the nanomaterials. The conjugation of Tunicamycin to Au NPs as well as the peptide nanotubes was confirmed by Fourier Transformation Infrared (FTIR) spectroscopy, absorbance spectroscopy, transmission electron microscopy (TEM), and atomic force microscopy (AFM). Decreased viability of capillary endothelial cells by these nano-formulations was confirmed by the 3-(4,5-methylthiazol-2-yl)-2,5-dipheyl-tetrazolium bromide (MTT) assay. Down-regulation of CDK4 and phospho-Rb (Ser249/Thr252) expression indicated slowing down of the cell cycle in G1. On the other hand, up-regulated expression of GRP-78 and those of its down-stream transducers IRE-1 and PERK/phospo-PERK not only confirmed the presence of “ER stress” but also the activation of the upr-mediated cellular event. In fact, translational attenuation of mannosylphospho dolichol synthase (DPMS) indeed confirmed the activation of upr-signaling. Down-regulation of caspase-9 and caspase-3 in cells treated with nano-formulated Tunicamycin may suggest a deviation from the central dogma of upr-signaling, whereas the “ER stress”-induced upr-mediated cell death suggests the presence of a non-canonical pathway.
Materials and methods
Materials-Hydroxyurea, dimethylsulfoxide (DMSO), nystatin, anti-phosphoserine monoclonal antibody, ethylenediamine tetra acetic acid (EDTA, sodium salt), Tunicamycin, MTT, and antibody to β-actin (mouse monoclonal) were obtained from Sigma Aldrich (St. Louis, MO). Rabbit polyclonal antibody for GRP-78 was from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-IRE1 (rabbit polyclonal) antibody, and antibody to phospho PERK were from Abcam (Cambridge, MA); antibodies to p53, phospho p53 (ser392), cyclin D1, and CDK4 were from BD Bioscience, (San Jose, CA). Anti-caspase 3 & 9 (mouse monoclonal) and anti-phospho-Rb (Ser249/Thr252; mouse monoclonal) antibodies were from Oncogene (San Diego, CA). Anti-Rb (mouse monoclonal) antibody was from EMD Biosciences (La Jolla, CA). HRP-conjugated goat anti-rabbit IgG/anti-mouse IgG, streptavidin and ECL chemiluminescence detection kit were from GE Healthcare (Piscataway, NJ). Biotinylated protein molecular weight markers and all electrophoresis reagents were obtained from Bio-Rad Laboratories (Hercules, CA). All other chemicals and solvents used were of the highest purity available. The cell culture wares were from Sarstedt (Newton, NC) and fetal bovine serum was purchased from HyClone Laboratories (Logan, UT). The nanoparticles of various sizes were either synthesized in the laboratory or purchased from Electron Microscopy Sciences (Hatfield, PA).
Synthesis and self-assembly of nanotubes
The bolaamphiphile bis (N-α-amido-threonine) 1,3-propane dicarboxylate contains threonine head groups, which are connected by a propyl carbon chain and was synthesized according to previously established methods (16). The intermediate obtained was washed with cold citric acid and sodium bicarbonate and recrystallized from dimethyl formamide (DMF). The product obtained was washed with cold acetone, and recrystallized from methanol. Individual stock solutions (8 mM) of the bis (N-alpha-amido-threonine)-1,3 propane dicarboxylate monomers were prepared in buffer solution of pH 5. In general, the materials were allowed to self-assemble for 7-10 days. Aggregates of nanotubular assemblies formed were then washed in nanopure water, followed by sonication for 30 min. The sizes and morphologies of the structures formed were examined by TEM, SEM, and AFM. The formed nanotubes were then used for binding to Tunicamycin either by adsorption or by covalent binding between the carboxyl groups of the nanotubes and the –OH groups of Tunicamycin. The attachment of Tunicamycin to the microtubes was confirmed by FTIR spectroscopy, TEM, and absorbance spectroscopy.
Attachment of Tunicamycin to the self-assembled nanotubes
The self-assembled nanotubes possess free carboxylic groups, which can be chemically modified. For binding Tunicamycin to the nanotubes, we conducted a selective esterification reaction between the primary alcohol group of Tunicamycin and the carboxylic acid groups of the nanotubes. Selective esterification of primary alcohols in the presence of secondary alcohols requires the use of specific catalytic agents. It has been shown that the use of catalysts such as Hf(IV)4 or Zr(IV)4 salts or 2,4,6-Trimethylpyridine have been efficient in selective esterification of primary alcohols (47-49), primarily due to the difference in the reactivity of the primary and secondary alcohol groups. Tunicamycin possesses a single primary alcohol group and seven secondary alcohol groups. In order to selectively bind the primary alcohol group of Tunicamycin to the nanotubes, without causing any major change to the other functional groups, the nanotubes were first dried at 50 °C overnight under nitrogen to remove any residual water. The dried nanotubes were then treated with thionyl chloride in the presence of triethylamine to convert the carboxyl acid groups to the corresponding acid chlorides. The product obtained was centrifuged. The nanotubes were then allowed to react with Tunicamycin (1.0 mmol) in the presence of 2,4,6-Trimethylpyridine (1.5 eq) in DMSO for 2 hours at 20 °C. The formed products were centrifuged and washed thoroughly with nanopure water to remove any unreacted excess reagents. The esterified product was confirmed by FTIR spectroscopy for the formation of functionalized nanotubes.
Encapsulation of Tunicamycin within threonine based peptide nanotubes
The incorporation of Tunicamycin into the nanotubes was examined by incubating the drug with the nanotubes. Since, the threonine nanotubes possess free hydroxyl and carborxylic acid groups, it was expected that threonine nanotubes would interact with Tunicamycin via strong hydrogen bonding. Tunicamycin was encapsulated into nanotubes by incubating the drug with the nanotubes at 4 °C for 48 hours under mild agitation. The samples were then washed and centrifuged to remove the excess Tunicamycin not incorporated. The incorporation of Tunicamycin was confirmed by TEM and FTIR analyses.
Conjugation of Au NPs to Tunicamycin
The Au NPs (20 or 50 nm) were incubated with reduced lipoic acid, thus functionalizing the nanoparticles with the thiol groups, while the carboxylic groups would be free to react. The nanoparticles of various sizes were either synthesized in the laboratory (50) or purchased from Electron Microscopy Sciences. The nanoparticles were allowed to react with reduced lipoic acid in nanopure water for 24 hours at 4 °C under mild agitation and under nitrogen. The nanoparticles were then washed and centrifuged to remove any unbound lipoic acid. The incorporation of lipoic acid was confirmed by the shift in the absorbance spectrum observed for the nanoparticles. The functionalized nanoparticles were then treated with Tunicamycin in the presence of 2,4,6-Trimethylpyridine (1.5 eq) in DMSO for 3 hours at 20 °C, washed, centrifuged, and dialyzed using snake skin dialysis tubing to remove any unreacted products. The incorporation of Tunicamycin to the nanoparticles was confirmed by TEM.
Culturing of capillary endothelial cells
The capillary endothelial cells were from the laboratory stock of a non-transformed endothelial cell line from bovine adrenal medulla and maintained as previously described (51). Synchronized (45,52) cultures were treated with Tunicamycin nanoparticles (1 µg/mL) for 1 h in EMEM containing 2% fetal bovine serum (heat-inactivated) unless mentioned otherwise. To synchronize, the cells were seeded in EMEM containing 10% fetal bovine serum and antibiotic mixture (penicillin/streptomycin/fungizone/nystatin) for 24 hours at 37 °C in a humidified CO2 incubator (5% CO2 and 95% air). At the end, the media were removed; The cells were washed three times with PBS, pH 7.2 and incubated for 48 hours in serum-free EMEM with one-third amount of antibiotic mixture containing 2 mM hydroxyurea. After 48 hours, the media were removed, cells were washed three times with PBS, pH 7.2, and re-incubated in serum-free EMEM with one-third amount of antibiotic mixture for an additional 24 hours. The media were removed, the cells were washed ones with PBS, pH 7.2 and incubated with Tunicamycin nanoparticles in EMEM containing 2% fetal bovine serum (heat-inactivated). FACS analysis of the synchronized capillary endothelial cells has been published earlier and the details can be found in ref(45).
Assessment of cell viability
The capillary endothelial cells (1×104/well) were grown in 96 well plates in EMEM supplemented with 10% FBS for 24 hours. After synchronization, cells were treated with no Tunicamycin (control), or native Tunicamycin, or with Tunicamycin nanoparticles (conjugated to 20 or 50 nm gold particles) for 1 h at 37 °C in 100 µL EMEM containing 2% fetal bovine serum. The cell viability was measured by MTT assay (53). Briefly, at the end of the incubation, the media were removed and the cells were incubated with MTT (100 µg in 20 µL EMEM with 2% fetal bovine serum) for 3 h at 37 °C. Media were removed and 100 µL DMSO was added to each well. Absorbance of solubilized formazan formed intracellularly was measured at 595 nm using an ELISA reader (Model 250; Bio-Rad Laboratories, Hercules, CA).
SDS-PAGE and western blot analyses
Performed as before (26,54,55) with 7.5% gel concentration.
Statistical analysis
The statistical analysis was carried out with Graph Pad Prism 4 software (Graph Pad Software Inc., San Diego, CA). Quantitative data are presented as mean plus S.E. The mean ± S.E. was calculated by one-way analysis of variance (ANOVA). Significance between groups was further analyzed using the post hoc Tukey’s test.
Results
Synthesis of Au NPs conjugated to Tunicamycin
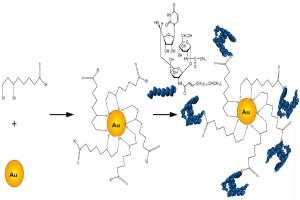
Au NPs were synthesized by using wet chemical methods (50). The average diameter of the Au NPs used for functionalization with Tunicamycin was 20 nm. The scheme for attachment of Tunicamycin to the Au NPs is shown in Figure 1.
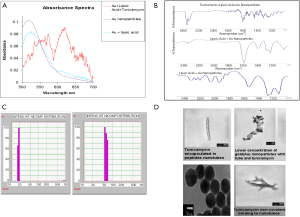
To functionalize the surface of the Au NPs, lipoic acid was first covalently bound to the nanoparticles and then treated with Tunicamycin. Figure 2A explains that there has been a shift in the absorbance from 515 to 524 nm upon binding to lipoic acid. Furthermore, FTIR spectroscopy indicates that upon treatment with Tunicamycin, there are two distinct peaks at 560 and 619 nm, respectively. A red shift to 560 nm indicates the attachment of Tunicamycin, while the peak at 619 nm is most likely due to the presence of aggregates formed during the reaction process (Figure 2B). Dynamic Light Scattering Studies of Au nanoparticles indicate that the Au NPs bound to Tunicamycin are indeed of 20 and 50 nm, respectively (Figure 2C). TEM has provided the morphology of various Tunicamycin nanoparticles, viz., Tunicamycin encapsulated in peptide nanotubes, bound of Au NPs to tubes and Tunicamycin, Au NPs bound to Tunicamycin nanotubes, etc. (Figure 2D).
Effect of Tunicamycin nanoparticles on cell viability
Synchronized capillary endothelial cells were incubated either with native Tunicamycin (1 µg/mL) or with 1 µg/mL of Tunicamycin nanoparticles for 1 h at 37 °C in a CO2 incubator (5% CO2 and 95% air) in 96-well microtiter plates. At the end, the plates were processed for the MTT Assay. The results (Figure 3A) indicate that the cell viability was reduced to almost 50% (P<0.001) when treated with Tunicamycin conjugated to 20 or 50 nm gold particles. Native Tunicamycin under similar condition had no effect. Importantly, both Tunicamycin nanotubes and nanoparticles were equally effective.
Tunicamycin nanoparticles inhibit cell cycle progression
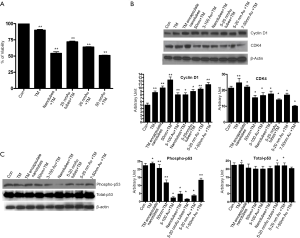
To evaluate the biochemical pathway(s) the Tunicamycin nanoparticles likely to interfere, we have analyzed their effect on the cell cycle progression. We have focused primarily on the D-type cyclins and their kinase partners, i.e., CDKs along with the Rb, a transcriptional initiator. Expression of cyclin D1, its catalytic partner, CDK4 and Rb was analyzed by western blotting. The expression of cyclin D1 was enhanced in cells treated with all nano-formulated Tunicamycin, and the native Tunicamycin (Figure 3B). On the other hand, the CDK4 expression was down regulated in all except in native Tunicamycin and Tunicamycin encapsulated nanotubes (Figure 3B). The logical conclusion is Tunicamycin impacted negatively on the cell cycle progression and most likely has caused a cell cycle arrest in G1.
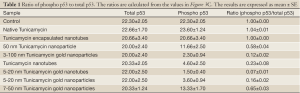
p53 is a transcription factor and often considered as the gate keeper for the cell cycle. Upon phosphorylation, p53 migrates to the nucleus and activates gene transcription. When analyzed by western blotting, the expression of phosphorylated p53 (i.e., phosphorylated in serine-392; p53pser392) was enhanced in cells treated with native Tunicamycin and Tunicamycin encapsulated nanotubes over the untreated control (Figure 3C). p53pser392 expression however was reduced ~50% in cells treated with 50 nm Tunicamycin nanoparticles, and 50 nm gold bound to Tunicamycin. The expression was much more reduced in cells treated with 100 nm gold bound to Tunicamycin, Tunicamycin nanotubes, 20 nm gold bound to Tunicamycin nanotubes and 20 nm gold bound to Tunicamycin, respectively. Total p53 was upregulated only in cells treated with either native Tunicamycin, or with 5-20 nm Tunicamycin gold nanotubes, or with 5-20 nm Tunicamycin gold nanoparticles (P<0.001). In all other cases, total p53 was down regulated non-significantly. If the ratio of phospho-p53 to total p53 is ~1.0 in native Tunicamycin treated cells then it is ~0.1-0.2 in cells treated with 5-20 nm Tunicamycin gold nanotubes, 3-100 nm Tunicamycin gold nanoparticles, Tunicamycin nanotubes, and 5-20 nm Tunicamycin gold nanoparticles, respectively. All others are in-between these values (Figure 3C, Table 1). It is therefore, concluded that nano-formulation of Tunicamycin inhibits phosphorylation of p53, which in turn may inhibit its translocation to the nucleus and gene transcription.
Full table
To evaluate if there is indeed a cell cycle inhibition we monitored the status of Rb, a cell cycle inhibitor protein. We have analyzed the expression of both total and phospho-Rb (Rb/pRb) by western blotting in cells treated with nano-formulated Tunicamycin. The results in Figure 4 explain that total Rb is down regulated (P<0.001) in cells treated with Tunicamycin encapsulated nanotubes, 3-100 nm Tunicamycin gold nanoparticles, and with Tunicamycin nanotubes. All other cases including the native Tunicamycin the changes were none or bare minimal. This was however different with the phospho-Rb (Ser249/Thr252) expression. In most cases the expression was reduced significantly (P<0.0001). The ratio of phospho-Rb to total Rb was reduced by ~34.4% to 64.2% (Table 2). This reduction was only 20% in cells treated with native Tunicamycin.
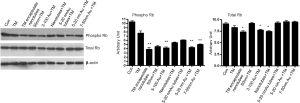
Full table
Tunicamycin Au NPs accelerate “ER stress”
It has been shown before that native Tunicamycin induces “ER stress” in capillary endothelial cells when treated for as early as 3 hours. We then compared the development of “ER stress” in a synchronized population of capillary endothelial cells treated with various nano-formulated Tunicamycin and native Tunicamycin just for 1 h. We have analyzed the GRP-78/Bip expression by western blotting as a quantitative measure of “ER stress”. Our results indicate that GRP-78/Bip expression was down regulated by ~28% in cells treated with native Tunicamycin, i.e., no detectable “ER stress”. The effect of 20 nm gold bound to Tunicamycin nanotubes was neutral. On the other hand, the GRP-78/Bip expression was increased by ~126-186% in cells treated with Tunicamycin nanotubes, and 20 or 50 nm gold bound to Tunicamycin, respectively over the control (Figure 5A).
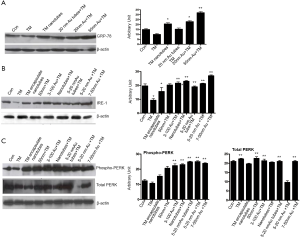
Tunicamycin Au NPs activates upr signaling
It has been observed recently that Tunicamycin inhibits angiogenesis in vitro and in vivo, and prevents the breast cancer progression in athymic nude mice by inducing “ER stress” in tumor microvasculature as well as in tumor cells (26). The above results support convincingly that Tunicamycin in its nano-formulation is highly active in inhibiting in vitro angiogenesis by blocking the cell cycle progression, and inducing the “ER stress”. Since, uncontrolled “ER stress” activates the upr signaling by attenuating the transcription and translation through IRE-1, ATF6/ATF4 (both are transcriptional attenuators) and PERK (a translational attenuator), we have analyzed here the expression of IRE-1 and PERK/phospho-PERK. The results (Figure 5B) indicate that the nano-formulation of Tunicamycin did not affect the expression of IRE-1. On the other hand, the PERK phosphorylation was activated significantly (P<0.0001) in cells treated with 3-100 nm gold Tunicamycin nanoparticles to 7-50 nm Tunicamycin Au NPs. Native Tunicamycin had no effect. The expression of total PERK however was increased in cells treated with native Tunicamycin, 50 nm Tunicamycin nanoparticles, 3-100 nm Tunicamycin gold nanoparicles, 5-20 nm Tunicamycin gold nanotubes, and 7-50 nm Tunicamycin Au NPs (Figure 5C). Interestingly, the ratio of phospho-PERK to total PERK revealed an increase of ~32.2% to 340.7% in all nano-formualted Tunicamycin used here (Table 3). The only exception however was the native Tunicamycin which exhibited ~18.6% reduction.
Full table
Tunicamycin Au NPs down-regulate DPMS expression
DPMS is an obligatory requirement for the synthesis of lipid-linked oligosaccharide (LLO; i.e., Glc3Man9GlcNAc2-PP-Dol), a pre-requisite for N-glycosylation (56,57). It is also an activator of GPT, the enzyme competitively inhibited by Tunicamycin (58) while DPMS itself is activated by phosphorylation (59). Furthermore, DPMS overexpression enhances capillary endothelial cell proliferation and accelerates the angiogenic process (55). Additionally, there is a cross-talk between the DPMS and GPT (60). Earlier, analysis of cell proteome by Raman Spectroscopy (C=O stretching) indicated a considerable amount of protein denaturation following Tunicamycin treatment (61). In order to evaluate the status of DPMS in cells treated with Tunicamycin nano-formulations, DPMS expression was monitored in treated cells by western blotting. The results (Figure 6) indicate that the expression of DPMS was reduced by ~33% when treated with native Tunicamycin. But, its expression was reduced by ~66-93% in cells treated with Tunicamycin nano-formulations. This correlates extremely well with upregulated phospho-PERK expression, a translational attenuator under “ER stress”. Therefore, decreased DPMS expression in cells treated with nano-formulated Tunicamycin is expected to be a significant contributing factor for the development of upr-mediated capillary endothelial cell death. The information thus confirms that nano-formulated Tunicamycin mediates its effect by inducing upr signaling.
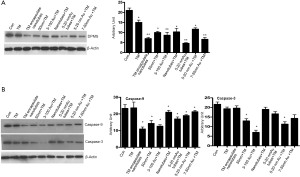
Tunicamycin nano-formulations down-regulate caspase-9 and caspase 3 expression
Activation of pro-caspase 9 to caspase-9 in the aptosome is the initiation of the apoptotic process and the process is concluded by the activation of caspase-3. The evidences presented so far have directed our attention towards the apoptotic death of the cells treated with Tunicamycin nano-formulations. But the information missing was the status of caspase-3 and caspse-9. Therefore, we have analyzed the expression of caspase-3 and caspase-9 by western blotting in cells exposed to all seven nano-formulations of Tunicamycin. The expression of caspase-9 was down regulated (P<0.001) in cells treated with all Tunicamycin nano-formulations with no change with native Tunicamycin (Figure 6B). It was selective in the case of caspase-3. The down regulation was markedly enhanced (P<0.001) with 50 nm Tunicamycin nanoparticles, 3-100 nm Tunicamycin gold nanoparticles, and 5-20 nm Tunicamycin Au NPs (Figure 6B).
Discussion
In asparagine-linked (N-linked) glycoproteins assembly, a core oligosaccharide unit consisting of fourteen sugar residues needs to be added to nascent proteins prior to its extensive modification by removal and addition of sugar residues in the endoplasmic reticulum (ER) and the Golgi complex (62). The modifications reflect a spectrum of glycoprotein functions and are best understood by blocking the availability of the core oligosaccharide unit (63-66). Using the best known N-glycosylation inhibitor, we have established in recent years that Tunicamycin (a natural product and a biologic) inhibited angiogenesis (i.e., capillary endothelial cell proliferation) in vitro and in athymic nude mice in vivo. It also prevented the progression of a double- and a triple-negative breast cancer in nude mice when administered intravenously or given orally (26). It is also fifteen times more potent that the currently FDA approved breast cancer therapeutic, Taxol. Tunicamycin is active only in G1 and its inhibitory action cannot be reversed either by VEGF, or by FGF2, or by high serum concentration. At the cellular level, Tunicamycin inhibits phospho tyrosine kinase activity and down-regulates the phospho VEGFR I & II levels, significantly even though Tunicamycin has no known tyrosine kinase inhibitory activity in vitro. More importantly, Tunicamycin has been found to be equally effective against neo-vascularization and the proliferation of a number of different human breast cancer cells. A breast cancer therapeutic of a dual action like Tunicamycin has not been discovered before. Therefore, our attempt to develop a nano-formulation of Tunicamycin has been a step forward and it has paid off. The nano-formulated Tunicamycin tested here has been found to be at least three times more potent than its native formulation.
The focus of our study has been to evaluate the efficacy of nano-formulated Tunicamycin over the native molecule. The question was, if nanoparticles are more effective than native Tunicamycin then a road map of the cells behavior while subjecting them to a shorter exposure could lead to a fundamental discovery. Therefore, 1 h exposure was quite reasonable, and based on our previous experience where we have seen cellular changes after exposing cells with native Tunicamycin for 3 h. Decreased cell viability correlated extremely well with the expression of cyclin D1 and CDK4 expression in cells treated with nano-formulated Tunicamycin. It is the presence of each component in the cyclin D1-CDK4 complex that matters most than the contribution of the individual component. In the present study, either (I) expression of both cyclin D1 and CDK4 were down regulated in some cases; or (II) in some cases when cyclin D1 expression was upregulated, the CDK4 expression was down-regulated and vice versa. This provides a strong indication for slowing down of the cell cycle with a possibility of its arrest in G1. The inhibition of cell cycle progression has been further substantiated with the down-regulated expression of phospho-Rb (Ser249/Thr252). p53 is expected to play a major role under such circumstance. Total p53 expression is significantly increased (P<0.001) in cells treated with 5-20 nm Tunicamycin gold nanotubes and 5-20 nm Tunicamycin gold nanoparticles. The rest of the nano-formulations had no effect (Figure 3C). On the other hand, the expression of phospho-p53 is down-regulated in all cells treated with Tunicamycin nano-formulation. The reasons may not be all that clear. Because, we have used p53 whose serine-392 is phosphorylated. It may not be the target responding adequately to the p53 level and/or may be degraded faster under the cellular environment created by the current experimental condition. Future monitoring of other phosphorelation target(s) is expected to clarify any anomalous behavior these cells may exhibit.
Increased GRP-78 expression indicates “ER stress”. High GRP-78 level in cells treated with Tunicamycin nanotubes, 20 nm Tunicamycin gold nanoparticles, and 50 nm Tunicamycin Au NPs establishes “ER stress”. Most interesting, however that GRP-78 expression is down-regulated in cells treated with equal amount (i.e., 1 µg) of native Tunicamycin. This strongly suggests that nano-formulated Tunicamycin is at least three times more potent. GRP-78 transduces “ER stress” through IRE-1, ATF6/ATF4 and PERK, components of upr signaling for apoptotic cell death. In this study we have examined the status of IRE-1, total PERK and phospho-PERK in cells treated with nano-formulated Tunicamycin. Both IRE-1 and phospho-PERK are elevated (P<0.0001; Figure 5B,C) in most cases treated with nano-formulated Tunicamycin whereas these are down regulated in cells treated with native Tunicamycin (P<0.001). The ratio of phospho-PERK to total PERK is presented in Table 3. Phospho-PERK is down regulated only in cells treated with native Tunicamycin, whereas in all nano-formulated the ratio has been increased. If the ratio is 1.0 in cells exposed to native Tunicamycin then it is between 1.6-5.5 fold higher in cells exposed to nano-formulated Tunicamycin. Information on ATF6/ATF4 could have added another segment of the upr signaling without contributing significantly to the net effect of what we have already observed. For example, the down regulated expression of DPMS. PERK is a translational attenuator and its activation through phosphorylation inhibits protein synthesis by phosphorylating the translation initiator eIf2. Therefore, DPMS down regulation with increasing PERK phophorylation satisfied the criteria for “ER stress”-induced upr-mediated apoptotic death. Our results on down-regulated expression of caspase-9 and consequently that of caspase-3 challenge the current hypothesis. Considering these findings we suggest: (I) the induction of “ER stress” and the upr-mediated apoptosis is a non-linear process; (II) There may exist a non-canonical pathway of cell death when the “ER stress” is induced; and (III) there is a possibility of autophagy and/or necrotic death. Obviously, more work is warranted to answer these questions. In addition, it would be interesting to know if nano-formulated Tunicamycin follows a single set of signaling pathways or differs based on the nature of the formulation.
It has been proposed earlier that native Tunicamycin enhanced the radio-sensitivity of U251 glioma and BXPC3 pancreatic adenocarcinoma cells (67), and also a combination of Tunicamycin with anticancer drugs [actinomycin D, vincristine (VCR), cis-diaminedichloroplatinum (II) (CDDP) and DXR] synergistically enhances their toxicity in multidrug-resistant human ovarian cystadenocarcinoma cells (68). This opens the door for developing Tunicamycin as a combination drug & radio therapy, and also the inherent property of Tunicamycin would make it more attractive to developing imaging tools.
Acknowledgments
The authors greatly appreciate the help from Jesus A. Santiago, an inter-campus graduate student during the course of this study.
Funding: The work was supported in part by the grant from Susan G Komen for Cure BCRT0600532.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Rao V. L. Papineni, Pataje G.S. Prasanna, Mansoor M. Ahmed) for the series “Nanotechnology in Radiation Research” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.3978/j.issn.2218-676X.2013.08.16). The series “Nanotechnology in Radiation Research” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All animal experiments were approved by the animal ethics committee and all operations complied with the animal experiment regulations.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Dreaden EC, El-Sayed MA. Detecting and destroying cancer cells in more than one way with noble metals and different confinement properties on the nanoscale. Acc Chem Res 2012;45:1854-65. [PubMed]
- Karimi Z, Karimi L, Shokrollahi H. Nano-magnetic particles used in biomedicine: core and coating materials. Mater Sci Eng C Mater Biol Appl 2013;33:2465-75. [PubMed]
- Nakatsuka N, Barnaby SN, Tsiola A, et al. Self-assembling peptide assemblies bound to ZnS nanoparticles and their interactions with mammalian cells. Colloids Surf B Biointerfaces 2013;103:405-15. [PubMed]
- de la Rica R, Matsui H. Applications of peptide and protein-based materials in bionanotechnology. Chem Soc Rev 2010;39:3499-509. [PubMed]
- Luo YL, Shiao YS, Huang YF. Release of photoactivatable drugs from plasmonic nanoparticles for targeted cancer therapy. ACS Nano 2011;5:7796-804. [PubMed]
- Cheng Y, Meyers JD, Broome AM, et al. Deep penetration of a PDT drug into tumors by noncovalent drug-gold nanoparticle conjugates. J Am Chem Soc 2011;133:2583-91. [PubMed]
- Abdullah-Al-Nahain. Development of disulfide core-crosslinked pluronic nanoparticles as an effective anticancer-drug-delivery system. Macromol Biosci 2011;11:1264-71. [PubMed]
- Corbierre MK, Cameron NS, Sutton M, et al. Gold nanoparticle/polymer nanocomposites: dispersion of nanoparticles as a function of capping agent molecular weight and grafting density. Langmuir 2005;21:6063-72. [PubMed]
- Wang Z, Jia L, Li MH. Gold nanoparticles decorated by amphiphilic block copolymer as efficient system for drug delivery. J Biomed Nanotechnol 2013;9:61-8. [PubMed]
- Aryal S, Pilla S, Gong S. Multifunctional nano-micelles formed by amphiphilic gold-polycaprolactone-methoxy poly(ethylene glycol) (Au-PCL-MPEG) nanoparticles for potential drug delivery applications. J Nanosci Nanotechnol 2009;9:5701-8. [PubMed]
- Song Q, Yao L, Huang M, et al. Mechanisms of transcellular transport of wheat germ agglutinin-functionalized polymeric nanoparticles in Caco-2 cells. Biomaterials 2012;33:6769-82. [PubMed]
- Huang M, Fong CW, Khor E, et al. Transfection efficiency of chitosan vectors: effect of polymer molecular weight and degree of deacetylation. J Control Release 2005;106:391-406. [PubMed]
- Dehsorkhi A, Hamley IW, Seitsonen J, et al. Tuning self-assembled nanostructures through enzymatic degradation of a peptide amphiphile. Langmuir 2013;29:6665-72. [PubMed]
- Hashida Y, Umeyama T, Mihara J, et al. Development of a novel composite material with carbon nanotubes assisted by self-assembled peptides designed in conjunction with β-sheet formation. J Pharm Sci 2012;101:3398-412. [PubMed]
- Sidelman N, Rosenberg Y, Richter S. Peptide-based spherulitic films--formation and properties. J Colloid Interface Sci 2010;343:387-91. [PubMed]
- Johnson KT, Gribb TE, Smoak EM, et al. Self-assembled nanofibers from leucine derived amphiphiles as nanoreactors for growth of ZnO nanoparticles. Chem Commun (Camb) 2010;46:1757-9. [PubMed]
- Smoak EM, Carlo AD, Fowles CC, et al. Self-assembly of gibberellic amide assemblies and their applications in the growth and fabrication of ordered gold nanoparticles. Nanotechnology 2010;21:025603 [PubMed]
- Huang CC, Lo YW, Kuo WS, et al. Facile preparation of self-assembled hydrogel-like GdPO4*H2O nanorods. Langmuir 2008;24:8309-13. [PubMed]
- Ghadiri MR, Granja JR, Buehler LK. Artificial transmembrane ion channels from self-assembling peptide nanotubes. Nature 1994;369:301-4. [PubMed]
- Pandit A, Fay N, Bordes L, et al. Self-assembly of the octapeptide lanreotide and lanreotide-based derivatives: the role of the aromatic residues. J Pept Sci 2008;14:66-75. [PubMed]
- Gudlur S, Sukthankar P, Gao J, et al. Peptide nanovesicles formed by the self-assembly of branched amphiphilic peptides. PLoS One 2012;7:e45374 [PubMed]
- Petrov A, Audette GF. Peptide and protein-based nanotubes for nanobiotechnology. Wiley Interdiscip Rev Nanomed Nanobiotechnol 2012;4:575-85. [PubMed]
- Brea RJ, Reiriz C, Granja JR. Towards functional bionanomaterials based on self-assembling cyclic peptide nanotubes. Chem Soc Rev 2010;39:1448-56. [PubMed]
- Butler JS, Sadler PJ. Targeted delivery of platinum-based anticancer complexes. Curr Opin Chem Biol 2013;17:175-88. [PubMed]
- Seabra AB, Durán N. Biological applications of peptides nanotubes: an overview. Peptides 2013;39:47-54. [PubMed]
- Banerjee A, Lang JY, Hung MC, et al. Unfolded protein response is required in nu/nu mice microvasculature for treating breast tumor with tunicamycin. J Biol Chem 2011;286:29127-38. [PubMed]
- Schneider BP, Miller KD. Angiogenesis of breast cancer. J Clin Oncol 2005;23:1782-90. [PubMed]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000;100:57-70. [PubMed]
- Folkman J. Angiogenesis and breast cancer. J Clin Oncol 1994;12:441-3. [PubMed]
- Uhr JW, Scheuermann RH, Street NE, et al. Cancer dormancy: opportunities for new therapeutic approaches. Nat Med 1997;3:505-9. [PubMed]
- Gastl G, Hermann T, Steurer M, et al. Angiogenesis as a target for tumor treatment. Oncology 1997;54:177-84. [PubMed]
- Folkman J. The role of angiogenesis in tumor growth. Semin Cancer Biol 1992;3:65-71. [PubMed]
- Dass CR, Tran TM, Choong PF. Angiogenesis inhibitors and the need for anti-angiogenic therapeutics. J Dent Res 2007;86:927-36. [PubMed]
- Dai J, Rabie AB. VEGF: an essential mediator of both angiogenesis and endochondral ossification. J Dent Res 2007;86:937-50. [PubMed]
- Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2006. CA Cancer J Clin 2006;56:106-30. [PubMed]
- Folkman J. What is the evidence that tumors are angiogenesis dependent? J Natl Cancer Inst 1990;82:4-6. [PubMed]
- Banerjee DK. Microenvironment of endothelial cell growth and regulation of protein N-glycosylation. Indian J Biochem Biophys 1988;25:8-13. [PubMed]
- Oliveira CM, Banerjee DK. Role of extracellular signaling on endothelial cell proliferation and protein N-glycosylation. J Cell Physiol 1990;144:467-72. [PubMed]
- Tiganis T, Leaver DD, Ham K, et al. Functional and morphological changes induced by tunicamycin in dividing and confluent endothelial cells. Exp Cell Res 1992;198:191-200. [PubMed]
- Nguyen M, Folkman J, Bischoff J. 1-Deoxymannojirimycin inhibits capillary tube formation in vitro. Analysis of N-linked oligosaccharides in bovine capillary endothelial cells. J Biol Chem 1992;267:26157-65. [PubMed]
- Banerjee DK, Vendrell-Ramos M. Is asparagine-linked protein glycosylation an obligatory requirement for angiogenesis? Indian J Biochem Biophys 1993;30:389-94. [PubMed]
- Nguyen M, Strubel NA, Bischoff J. A role for sialyl Lewis-X/A glycoconjugates in capillary morphogenesis. Nature 1993;365:267-9. [PubMed]
- Pili R, Chang J, Partis RA, et al. The alpha-glucosidase I inhibitor castanospermine alters endothelial cell glycosylation, prevents angiogenesis, and inhibits tumor growth. Cancer Res 1995;55:2920-6. [PubMed]
- Ingber DE, Folkman J. How does extracellular matrix control capillary morphogenesis? Cell 1989;58:803-5. [PubMed]
- Martínez JA, Torres-Negrón I, Amigó LA, et al. Expression of Glc3Man9GlcNAc2-PP-Dol is a prerequisite for capillary endothelial cell proliferation. Cell Mol Biol (Noisy-le-grand) 1999;45:137-52. [PubMed]
- Wang X, Zhen X, Wang J, et al. Doxorubicin delivery to 3D multicellular spheroids and tumors based on boronic acid-rich chitosan nanoparticles. Biomaterials 2013;34:4667-79. [PubMed]
- Yamamoto N, Obora Y, Ishii Y. Iridium-catalyzed oxidative methyl esterification of primary alcohols and diols with methanol. J Org Chem 2011;76:2937-41. [PubMed]
- Ishihara K, Kubota M, Kurihara H, et al. Scandium Trifluoromethanesulfonate as an Extremely Active Lewis Acid Catalyst in Acylation of Alcohols with Acid Anhydrides and Mixed Anhydrides. J Org Chem 1996;61:4560-7. [PubMed]
- Ishihara K, Nakayama M, Ohara S, et al. Direct ester condensation from a 1:1 mixture of carboxylic acids and alcohols catalyzed by hafnium(IV) or zirconium(IV) salts. Tetrahedron 2002;58:8179-88.
- Enustun BV, Turkevich J. Coagulation of colloidal gold. J Am Chem Soc 1963;85:3317-28.
- Banerjee DK, Ornberg RL, Youdim MB, et al. Endothelial cells from bovine adrenal medulla develop capillary-like growth patterns in culture. Proc Natl Acad Sci U S A 1985;82:4702-6. [PubMed]
- Martínez JA, Tavárez JJ, Oliveira CM, et al. Potentiation of angiogenic switch in capillary endothelial cells by cAMP: A cross-talk between up-regulated LLO biosynthesis and the HSP-70 expression. Glycoconj J 2006;23:209-20. [PubMed]
- Majumdar KN, Banerjee A, Ratha J, et al. Leishmanial lipid suppresses tumor necrosis factor alpha, interleukin-1beta, and nitric oxide production by adherent synovial fluid mononuclear cells in rheumatoid arthritis patients and induces apoptosis through the mitochondrial-mediated pathway. Arthritis Rheum 2008;58:696-706. [PubMed]
- Baksi K, Tavárez-Pagán JJ, Martínez JA, et al. Unique structural motif supports mannosylphospho dolichol synthase: an important angiogenesis regulator. Curr Drug Targets 2008;9:262-71. [PubMed]
- Zhang Z, Banerjee A, Baksi K, et al. Mannosylphosphodolichol synthase overexpression supports angiogenesis. Biocatal Biotransformation 2010;28:90-8. [PubMed]
- Banerjee DK, Scher MG, Waechter CJ. Amphomycin: effect of the lipopeptide antibiotic on the glycosylation and extraction of dolichyl monophosphate in calf brain membranes. Biochemistry 1981;20:1561-8. [PubMed]
- Chapman A, Trowbridge IS, Hyman R, et al. Structure of the lipid-linked oligosaccharides that accumulate in class E Thy-1-negative mutant lymphomas. Cell 1979;17:509-15. [PubMed]
- Kean EL. Site of stimulation by mannosyl-P-dolichol of GlcNAc-lipid formation by microsomes of embryonic chick retina. Glycoconj J 1996;13:675-80. [PubMed]
- Banerjee DK, Kousvelari EE, Baum BJ. cAMP-mediated protein phosphorylation of microsomal membranes increases mannosylphosphodolichol synthase activity. Proc Natl Acad Sci U S A 1987;84:6389-93. [PubMed]
- Banerjee DK. N-glycans in cell survival and death: cross-talk between glycosyltransferases. Biochim Biophys Acta 2012;1820:1338-46.
- Longas MO, Kotapati A, Prasad KP, et al. Balancing life with glycoconjugates: monitoring unfolded protein response-mediated anti-angiogenic action of tunicamycin by Raman Spectroscopy. Pure Appl Chem 2012;84:1907-18. [PubMed]
- Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem 2004;73:1019-49. [PubMed]
- Bernales S, Papa FR, Walter P. Intracellular signaling by the unfolded protein response. Annu Rev Cell Dev Biol 2006;22:487-508. [PubMed]
- Schröder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem 2005;74:739-89. [PubMed]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 2007;8:519-29. [PubMed]
- Lai E, Teodoro T, Volchuk A. Endoplasmic reticulum stress: signaling the unfolded protein response. Physiology (Bethesda) 2007;22:193-201. [PubMed]
- Contessa JN, Bhojani MS, Freeze HH, et al. Inhibition of N-linked glycosylation disrupts receptor tyrosine kinase signaling in tumor cells. Cancer Res 2008;68:3803-9. [PubMed]
- Hiss DC, Gabriels GA, Folb PI. Combination of tunicamycin with anticancer drugs synergistically enhances their toxicity in multidrug-resistant human ovarian cystadenocarcinoma cells. Cancer Cell Int 2007;7:5. [PubMed]