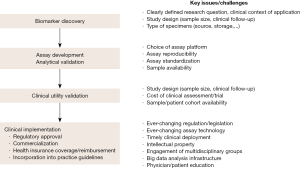
Cancer biomarker discovery and validation
Introduction
The Precision Medicine Initiative unveiled in January 2015, included an investment of $70 million to the National Cancer Institute (NCI), to “scale up efforts to identify genomic drivers in cancer and apply that knowledge in the development of more effective approaches to cancer treatment” (1). In the field of cancer research and care, the concept of precision medicine—prevention and treatment strategies that take individual variability into account—hinges on the development of valid biomarkers interrogating key aberrant pathways potentially targetable with molecular targeted or immunologic therapies (1). Although biomarkers such as prostate-specific antigen (PSA), have been known and used for decades to attempt to guide prognostic and therapeutic decisions, the recent revolution in molecular biology, with the rise of high-throughput sequencing and increased molecular characterization of tumor tissue has led to an exponential increase in attempts to measure and target aberrant pathways at the molecular level. Nevertheless, there has been a large gap between multiple initial reports of biomarkers, often with diagnostic performance that cannot be reproduced in later studies, and full clinical implementation and validation of the biomarkers due to issues in study design, assay platforms, and availability of specimens for biomarker development (2,3).
Nevertheless, with the recent emergence of highly selective molecular targeted agents and high-throughput genomic characterization technologies, robust and well-validated cancer biomarkers are increasingly needed. For instance, more than 90% of oncological drugs that enter clinical development will not reach market approval due to failure of clinical trials to demonstrate therapeutic benefit, contributing to costly and slow cancer drug development (4). As acknowledged by the USA Food and Drug Administration (FDA), the judicious use of biomarkers is expected to play an important role in minimizing risk of clinical trial failure by enriching the trial populations with specific molecular subtypes responding better to tested therapies. In this review, we overview recent trends in cancer biomarker development and discuss the issues in clinical translation of cancer biomarkers.
Biomarkers in cancer care
A biomarker is an objectively measured characteristic that describes a normal or abnormal biological state in an organism by analyzing biomolecules such as DNA, RNA, protein, peptide, and biomolecule chemical modifications (5). However, it must be acknowledged that the definition of biomarkers has been evolving over the past decade, with one especially broad definition by the World Health Organization suggesting that “A biomarker is any substance, structure or process that can be measured in the body or its products and influence or predict the incidence of outcome or disease.” (6,7). More specifically in terms of clinical utility, a cancer biomarker may measure the risk of developing cancer in a specific tissue or, alternatively, may measure risk of cancer progression or potential response to therapy. Besides providing useful information in guiding clinical decision making, cancer biomarkers are increasingly linked to specific molecular pathway deregulations and/or cancer pathogenesis to justify application of certain therapeutic/interventional strategies. The conceptual framework of cancer biomarker development has also been evolving with the rapid expansion of our omics analysis capability of clinical biospecimens based on the traditional path of biomarker deployment (5).
Cancer biomarkers can be classified into the following categories based on their usage. Predictive biomarkers predict response to specific therapeutic interventions such as positivity/activation of HER2 that predicts response to trastuzumab in breast cancer (8-10). Similarly, KRAS-activating mutations predict resistance to epidermal growth factor receptor (EGFR) inhibitors such as cetuximab in colorectal cancer (11). Prognostic biomarker, on the other hand, may not be directly linked to or trigger specific therapeutic decisions, but aim to inform physicians regarding the risk of clinical outcomes such as cancer recurrence or disease progression in the future. An example of a prognostic cancer biomarker is the 21-gene recurrence score which was predictive of breast cancer recurrence and overall survival in node-negative, tamoxifen-treated breast cancer (12). Another class of biomarker, the diagnostic biomarker, is used to identify whether a patient has a specific disease condition. Diagnostic biomarkers have recently been implemented for colorectal cancer surveillance by testing for stool cancer DNA (13).
Processes of biomarker development
Biomarker development involves multiple processes, linking initial discovery in basic studies, validation, and clinical implementation (Figure 1) (5,14-21). The ultimate goal of the processes is to establish clinically accessible biomarker tests with clinical utility, informing clinical decision-making to improve patient outcomes (21,22). However, there are many hurdles as evidenced by the low estimated rate (0.1%) of successful clinical translation of biomarkers (23). Here we elaborate each of the processes, which should be designed/planned prior to the conduct of the study to ascertain validity of cancer biomarkers.
Biomarker discovery
At the start of any biomarker development, biomarkers should be “discovered” and are typically validated within the same initial report. Validation based on predefined prediction rule in an independent patient series is ideal, but it is often substituted by cross-validation-based methods when independent patient sets are not available. The research question and plan, including the fundamental use of the biomarker, should traditionally be clearly defined prior to the analysis, although this can be challenging at the very early stages of biomarker development. In this era of ever-evolving high-throughput omics technologies where thousands of individual molecules can be easily interrogated without a priori assumptions, research hypotheses are often generated in a post hoc manner, following often serendipitous discovery from unbiased mining of the genome-wide measurements (data-driven hypothesis generation) (20). Another relevant issue to be addressed early in biomarker development is the target population to be tested in specific clinical contexts, which will guide subsequent clinical evaluation and implementation. In general, broader target populations could lead to increased costs and risks of failure during the development stage.
Study design/setting, from which analyzed biospecimens are derived, is the major source of bias that hampers subsequent biomarker development. Ideally, the specimens should be prospectively collected based on well-defined inclusion and exclusion criteria together with accompanying clinical annotations pre-specified in the study protocol. A cohort or case-control study design is typically employed. In a cohort study, clinical characteristics of enrolled individuals as well as information of intervention and follow-up are critical in identifying molecular correlates associated with clinical outcomes of interest. In a case-control study, potential confounding factors should be properly matched between cases and controls to minimize false discovery. In practice, biomarker discovery is often based on “samples of convenience”, which were incidentally available to the investigator at the time of research and collected without prior intention of specific biomarker discovery (24). This could introduce unrecognized confounding factors, which may contribute to the false positive associations of the biomarkers. The study design quality may be semi-quantitatively evaluated by using scores such as level of evidence scale proposed by Simon et al. (16). In general, evidence derived from large-scale well-predefined prospective trials is regarded as most reliable. Retrospective, observational studies may be affected by multiple sources of bias, which can be better identified if reporting guidelines such as Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK) for prognostic studies (25), Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) for observational studies (26) and Standards for Reporting of Diagnostic Accuracy (STARD) for diagnostic studies (27) are used to determine reliability and quality of biomarkers in the initial reports.
A common cause of failure in developing robust predictive and especially prognostic biomarkers is to define them based on clinically invalid surrogate endpoints such as objective response in oncology trials as well as short-term outcomes from retrospective studies. Biomarkers trained for poorly-defined endpoints are more likely to fail in subsequent prospective evaluation. A prognostic gene-expression signature trained on long-term outcome using archived specimens has been successfully validated in a series of independent clinical and experimental studies (28-31). While the most optimal setting is prospective sample collection and follow-up based on a fully predefined protocol, this requires costly and lengthy biomarker assessment, which hampers timely deployment of cancer biomarkers. As an alternative, retrospective analysis of samples archived as part of previously completed prospective trials (prospective-retrospective design) is proposed to shorten the time frame while ascertaining quality of study design (16). Another solution is to develop a biobank in which biospecimens and complete clinical annotations are prospectively accumulated based on well-defined protocols. However, in part due to the complex and heterogeneous nature of cancer, it has become increasingly recognized, that there is a need for larger integrated biobanks (32,33) which require careful development and adherence to published biobanking guidelines (34). The practical challenges of biobanking in cancer patients has been underlined by a recent USA survey of NCI-funded cancer researchers who conduct tissue-based research showing that 39-47% reported difficulty obtaining biospecimens of adequate numbers and quality and low-quality biospecimens resulted in 60% questioning their findings and 81% limiting the scope of their work (35). Quality of clinical annotations is another key factor in utilizing the resources to identify reliable biomarkers and validate their clinical utility. A recent NCI joint workshop recommended improved sharing of existing specimens and data and creation of NCI-wide inventory of prediagnostic specimens and cancer diagnosis data, ongoing engagement of the clinical, translational and basic research communities, and encouraging the development of pilot projects (18).
Robustness of sample processing and data analysis procedures is another factor that influences reproducibility of biomarker studies. For example, a high diagnostic accuracy of a peptide signature for ovarian cancer was not confirmed in subsequent independent reanalysis of the original dataset possibly due to variation in sample processing (36,37). One report of proteomic biomarker discovery noted that common statistical algorithms run on data with low sample sizes can overfit and yield misleading misclassification rates and that prefiltering variables exacerbated this problem (38). Similarly, a critical review of prognostic microarray studies in cancer revealed that half of the reported prognostic gene signatures were not reproducible due to critical flaws in the data analysis methods (39). These reports highlight the importance of careful assessment of technical soundness and methodological validity and disclosure of information to the research community to enable fair evaluation of reported biomarkers and identification of candidates for further development. In addition, ensuring reproducibility of bioinformatics analysis is a critical determinant of successful clinical translation of genome-based biomarkers. There have been several efforts to develop informatics infrastructure to address this issue, including public repository of datasets with relevant annotations on biological, clinical, and experimental parameters, analysis software repository, and systems to record whole process of data analysis itself to allow anyone to rerun or modify the analysis to verify robustness of reported findings (40,41).
Biomarker assay development and analytical validation
Following the discovery phase that typically includes internal validation, candidate biomarkers are adapted to clinically applicable assay platforms, and subjected to two types of validation, namely analytical validation, i.e., how accurately and reliably does the test measure the analyte(s) of interest in the patient specimen and clinical validation, i.e., how robustly and reliably is the test result correlated with the clinical phenotype or the outcome of interest. Analytical validation is typically performed by assaying the same set of samples by both the assay used in the initial discovery and the clinical deployment platform to determine robustness and reproducibility of the measurements. Frequently used assay technologies generally used for analysis of single gene/protein anomalies include real-time polymerase chain reaction (RT-PCR) to assess gene expression or DNA mutations (e.g., BRAF V600E mutation in melanoma), fluorescent in situ hybridization (FISH) to assess DNA copy number or genetic translocation (e.g., HER2 amplification, BCR-ABL translocation), and immunohistochemistry (IHC) to assess protein expression and subcellular localization (e.g., estrogen receptor expression in breast cancer).
More recently, several multi-gene assays classified as in vitro diagnostic multivariate index assays (IVDMIA) have been introduced into clinic (13,42,43). The implementation of gene expression-based multi-gene assays has been a challenging task due to poorer reproducibility of the measurements (44). Currently available tests, such as MammaPrint (45) and Oncotype Dx (12), are performed in centralized laboratories to minimize technical variability. Emerging technology such as direct digital counting of transcripts without target amplification could enable more robust gene expression measurements reproducible across individual laboratories (46,47). Resequencing of a targeted panel of genes (disease-specific, exome, etc.) has been tested as another option (48), identifying somatic DNA mutations potentially driving cancer in nearly 2/3 of patients with lung adenocarcinomas and linking to molecular targeted therapy in 28% of patients (49). Clinical sequencing is a promising approach, but the interpretation and reporting of incidental findings from non-targeted sequencing is still being debated (50). In addition, high demand on data analysis, referred as the “$1,000 genomic test [but] $100,000 genomic analysis”, is another layer of challenge in sequencing-based approaches (51). Capability to analyze formalin-fixed, paraffin-embedded (FFPE) tissue samples greatly enhances general applicability of biomarker assays (52-54). Emergence of highly sensitive assays, e.g., single cell profiling, are expected to enable analysis of body fluid-derived specimens such as whole blood, plasma, serum, ascites, and urine to assess circulating microRNA, circulating DNA, and circulating tumor cells (CTCs)-derived biomolecules (55,56). These technologies are expected to achieve less-invasive assessment of molecular biomarkers (liquid biopsy) (55). Circulating tumor DNA was highly accurate in assessing mutation status of BRAF V600E mutation (100% specificity and sensitivity reported) and KRAS point mutations (>90% sensitivity and specificity) in subjects with metastatic colorectal cancer in one blinded prospective trial (57). Another report, assessing the role of CTCs, defined as 5 or more per 7.5 mL of whole blood in this study, in metastatic breast cancer, did not find an improvement in outcomes after changing therapy in case of persistently elevated CTCs but confirmed that CTCs were strongly prognostic for overall outcome (58). In addition to their role in diagnosis, circulating cell-free microRNAs are also being currently assessed as a predictive cancer biomarker with some encouraging preliminary reports (59,60).
Validation of clinical utility
After analytical validity is confirmed, the biomarker assay in the clinical deployment platform must be evaluated to confirm its performance in predicting or diagnosing the clinical phenotype or outcome of interest as demonstrated in the discovery and initial validation phase (5,21,61). Ideally, the biomarker should be evaluated in statistically well-powered prospective trials as performed in the TransATAC study for breast cancer recurrence prediction (62). However, it is realistically infeasible to test all candidate biomarkers in this manner due to financial constraints and/or limited availability of patient cohorts. Therefore, similar to the setting of biomarker discovery, the use of prospective-retrospective design and/or biobank/biorepository samples could be a potential alternative to overcome these obstacles. Clinical utility assessment could also include analysis of clinically meaningful outcome benefit, comparative effectiveness, cost-effectiveness of biomarker-guided clinical care, and assessment of alternatives and availability of the biomarker based on real-world clinical data or mathematical modeling (21,63).
Clinical implementation
An analytically and clinically validated biomarker assay is now ready for implementation in clinical care. This phase includes the following four key elements, which vary widely across regions: regulatory approval, commercialization, coverage by health insurance companies, and incorporation in clinical practice guidelines. In the USA, there are two paths for regulatory approval: in vitro diagnostic device (IVD) as commercial medical device with 510(k) clearance overseen by the FDA, and laboratory developed tests (LDT), home-grown assay developed and optimized at a diagnostic lab performing the test, which will likely be regulated by the FDA although current oversight is more limited (64). Clinical biomarker tests must be conducted in diagnostic laboratories certified for Clinical Laboratory Improvement Amendments (CLIA) and in accordance with state-specific regulations. Coverage by health insurance is critical for physicians to order the tests. Assignment of current procedural terminology (CPT) codes as well as incorporation into clinical practice guideline/recommendation supports payer’s decision. Centers for Medicare & Medicaid Services (CMS) classifies the tests into tier 1 (CPT code-assigned, commonly performed tests) and tier 2 (less commonly performed tests grouped by complexity). CMS defers pricing for new CPT codes to the local Medicare administrative contractors in a procedure known as “gapfill”, which causes delayed reimbursement for many biomarker tests (65). Post-marketing clinical utility validation will further support the use of biomarker tests, and may result in indication for additional diseases and/or clinical scenarios. Resources such as the National Comprehensive Cancer Network Biomarkers Compendium (66) are available to access the current recommendation for biomarkers in clinical guidelines (67).
Cancer biomarkers currently available in clinic
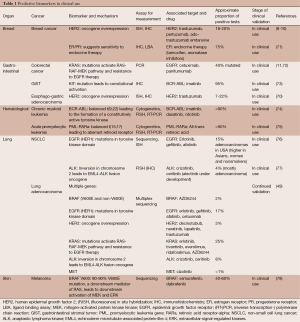
An example of a molecular biomarker in clinic is overexpression/amplification of HER2 (ERBB2), a member of the EGFR family, predictive of response to monoclonal antibodies such as trastuzumab and pertuzumab in breast cancer (8-10). It has been shown in pivotal phase III trials in breast cancer that subjects with HER2 overexpression (approximately 20% of patients) treated with anti-HER2 therapy have improved disease-free and overall survival (8-10). American Society of Clinical Oncology and College of American Pathologists recommend primarily IHC and in situ hybridization for assessment of HER2 status (68). Currently, the FDA has approved 10 HER2 assays as companion diagnostic devices (50% of all approved companion diagnostic devices) and 3 other HER2 assays as nucleic acid based tests cleared by the Center for Devices and Radiological Health [FDA website accessed on March 20th 2015 (69)]. HER2 overexpression is similarly predictive of response to trastuzumab in esophago-gastric adenocarcinoma (70). OmniSeq Target assay analyzes clinically actionable somatic DNA alterations in 23 known cancer-related genes, which acquired the New York state approval as LDT. Other major predictive biomarkers, including BCR-ABL in chronic myeloid leukemia and KRAS mutations in colorectal cancer and multiple mutations in non-small cell lung cancer (NSCLC), are listed in Table 1.
Full table
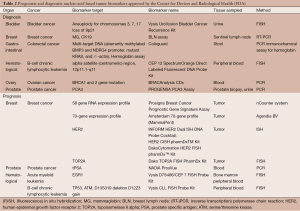
Despite the numerous prognostic biomarkers reported in the literature, only seven biomarkers have been approved by the FDA Center for Devices and Radiological Health (Table 2) (48). One of the major reasons is that prognostic prediction itself often does not directly change clinical decision making unless coupled to specific therapeutic options. Despite this, many other prognostic biomarkers are available through the LDT pathway. Mammaprint is one of the first gene expression signature-based assays based on the measurement of 70 genes to predict breast cancer recurrence after chemotherapy, which was recently adapted for use in FFPE tissue (45). Another gene expression-based assay, Oncotype Dx Breast Cancer Assay measures 21 genes predicting breast cancer recurrence in women with node negative or node positive, ER-positive, HER2-negative invasive breast cancer (12,79). Similar tests are also available for colon and prostate cancer, all of which analyze gene expression in tumor tissue (80,81). A 186-gene expression signature in non-tumor stromal liver tissue has been validated to predict hepatocellular carcinoma development and recurrence as well as liver cirrhosis progression, and was recently implemented in an FDA-approved diagnostic device (28-30).
Full table
Diagnostic biomarkers are one of the most diverse classes of biomarkers ranging from assays developed for cancer screening to diagnostic tests assessing progression of a known cancer (see Table 2 for a list of FDA-approved diagnostic genetic tests). One recent example of a diagnostic biomarker is Cologuard, a multigene DNA (KRAS mutations, aberrant NDRG4 and BMP3 methylation) stool test combined with fecal immunochemistry designed to screen for colorectal cancer in individuals at average risk of colorectal cancer. In a recent clinical trial of nearly 10,000 participants, sensitivity of the test for detecting colorectal cancer was higher than fecal immunochemical test alone (92.3% and 73.8% respectively) although the test also had a higher rate of false positives (specificity 86.6% and 96.4% for Cologuard and fecal immunochemical test respectively) (13). These encouraging results led to the approval of this test by the FDA in August 2014. Recently, there has also been increased interest in developing minimally invasive diagnostic tumor biomarkers, using the measurement of circulating DNA or microRNA. For instance, a new technology termed cancer personalized profiling by deep sequencing (CAPP-Seq) has been tested on circulating tumor DNA in patients with NSCLC. Levels of circulating DNA correlated with tumor volume and provided earlier response assessment than radiography in this preliminary trial while potentially allowing the non-invasive detection of actionable mutations (82). Another report, focusing on circulating microRNA serum profiles identified a microRNA profile thought to distinguish subjects with pancreatic cancer from healthy controls, even at early stages of the disease (83). This result requires further validation but may suggest a direction towards which the field of diagnostic biomarkers is moving. However, even when FDA-approved, commercialization may still be a challenge due to the high cost required for assay development.
Cancer biomarkers under evaluation in clinical trials
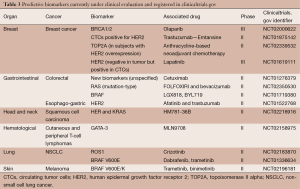
Multiple predictive biomarkers, mostly based on single gene/protein, are currently in phase II or III evaluation along with their companion therapeutic agents (Table 3). From this snapshot, the increasing importance of predictive biomarkers is apparent as is a trend to develop minimally invasive cancer biomarkers. Biomarkers validated in a certain type of cancer are undergoing discovery and validation in other cancers (for instance BRAF mutations or HER2 overexpression) underlining certain shared oncogenic drivers and less prevalent cancers are also benefitting from the rapid developments in the field. The 70-gene breast cancer signature is currently being evaluated for its recurrence-predictive capability in comparison to clinico-pathological assessment in a large prospective trial enrolling more than 6,600 subjects in nine countries (MINDACT study) with early results suggesting that the 70-gene signature added information to usual assessment (84).
Full table
Future perspectives and conclusions
In this review, we aimed to overview the current landscape of cancer biomarker development. The speed of technological development has highlighted the challenges facing regulatory oversight and legislation in their attempts to keep up with the rapid pace of scientific changes while allowing proper consideration to how the new biomarkers could shape the future of medicine (85,86). One of the major challenges is to manage the tradeoff between safety and speed of clinical translation. For example, regulation of LDT by the FDA will improve assay quality and safety and increase overall medical utility of the tests, while it could hamper timely deployment of the tests and benefit only large commercial laboratories with capabilities to accommodate the high requirements. The large amount of data generated by the assays have posed supplementary challenges in the analysis of “big data”, which requires massive computational resources for data storage, processing, and interpretation (87). Informatics resources such as ClinGen (88) are being developed to support the process. Also, systems to integrate genomic information with electronic medical records (EMRs) are actively developed, where protection of patient privacy is a central issue such as the Electronic Medical Records and Genomics Network (eMERGE), a NIH-funded consortium aiming to develop and disseminate approaches combining DNA biorepositories with EMRs (89). However, the integration of EMRs with genomic datasets remains in its infancy, due to a number of challenges including defining optimal storage standards of genomic data, integration of rich phenotype information, interpretation of complex data in a format easily accessible to clinicians and of course ethical, legal and social issues (90). Defining unified standard for the systems and data formats is particularly challenging due to the big financial/commercial interests.
Another crucial aspect of biomarker development, especially genomic biomarkers, is the issue of intellectual property. In the USA, a recent high profile Supreme Court decision, The Association for Molecular Pathology versus Myriad, determined that isolated but otherwise unmodified genes were products of nature and therefore not patent eligible subject matter (91). This decision was a response to an ongoing lawsuit between Myriad Genetics, who owned the exclusive rights to analyze the BRCA1 and BRCA2 gene mutations, and a coalition of groups who challenged the constitutionality and validity of the BRCA1 and BRCA2 gene patents. In this context, the USA Patent and Trademark Office has recently issued new guidelines which enforce more stringent criteria to patent natural products such as antibiotics, or even nucleic acids, peptides and proteins. These new guidelines have generated considerable concern in the biotechnology world due to their far-reaching consequences that are still being considered (92). Of note, genetic sequences are currently still patent-eligible in the European Union and in Australia if certain conditions are fulfilled (93,94). It is expected to take more time to reach a solution acceptable to all relevant parties.
Despite unclear future prospects and regulatory and legislative minefields, several examples of successful clinical translation summarized above have emphasized the challenges but also the opportunities at each step of cancer biomarker development. Acknowledging these challenges and implementing them in the design of biomarker development will help streamline the whole process, and eventually transform cancer patient care by fulfilling the vision of Precision Medicine.
Acknowledgments
Funding: This work was supported by the FLAGS foundation, the Nuovo-Soldati Cancer Research Foundation and an advanced training grant from Geneva University Hospital to NG and NIH/NIDDK R01 DK099558 and the Irma T. Hirschl Trust to YH.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Jian-Bing Fan) for the series “Application of Genomic Technologies in Cancer Research” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.3978/j.issn.2218-676X.2015.06.04). The series “Application of Genomic Technologies in Cancer Research” was commissioned by the editorial office without any funding or sponsorship. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med 2015;372:793-5. [PubMed]
- Tzoulaki I, Siontis KC, Ioannidis JP. Prognostic effect size of cardiovascular biomarkers in datasets from observational studies versus randomised trials: meta-epidemiology study. BMJ 2011;343:d6829. [PubMed]
- Ioannidis JP, Panagiotou OA. Comparison of effect sizes associated with biomarkers reported in highly cited individual articles and in subsequent meta-analyses. JAMA 2011;305:2200-10. [PubMed]
- Paul SM, Mytelka DS, Dunwiddie CT, et al. How to improve R&D productivity: the pharmaceutical industry's grand challenge. Nat Rev Drug Discov 2010;9:203-14. [PubMed]
- Committee on the Review of Omics-Based Tests for Predicting Patient Outcomes in Clinical Trials, Board on Health Care Services, Board on Health Sciences Policy, et al. Evolution of Translational Omics: Lessons Learned and the Path Forward. National Academies Press 2012.
- Lassere MN. The Biomarker-Surrogacy Evaluation Schema: a review of the biomarker-surrogate literature and a proposal for a criterion-based, quantitative, multidimensional hierarchical levels of evidence schema for evaluating the status of biomarkers as surrogate endpoints. Stat Methods Med Res 2008;17:303-40. [PubMed]
- World Health Organization. Biomarkers in risk assessment: Validity and validation. WHO 2001.
- Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med 2001;344:783-92. [PubMed]
- Romond EH, Perez EA, Bryant J, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med 2005;353:1673-84. [PubMed]
- Piccart-Gebhart MJ, Procter M, Leyland-Jones B, et al. Trastuzumab after adjuvant chemotherapy in HER2-positive breast cancer. N Engl J Med 2005;353:1659-72. [PubMed]
- Van Cutsem E, Köhne CH, Hitre E, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med 2009;360:1408-17. [PubMed]
- Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 2004;351:2817-26. [PubMed]
- Imperiale TF, Ransohoff DF, Itzkowitz SH. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med 2014;371:187-8. [PubMed]
- Simon R, Roychowdhury S. Implementing personalized cancer genomics in clinical trials. Nat Rev Drug Discov 2013;12:358-69. [PubMed]
- McShane LM, Cavenagh MM, Lively TG, et al. Criteria for the use of omics-based predictors in clinical trials. Nature 2013;502:317-20. [PubMed]
- Simon RM, Paik S, Hayes DF. Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst 2009;101:1446-52. [PubMed]
- Hayes DF. Biomarker validation and testing. Mol Oncol 2015;9:960-6. [PubMed]
- Schully SD, Carrick DM, Mechanic LE, et al. Leveraging biospecimen resources for discovery or validation of markers for early cancer detection. J Natl Cancer Inst 2015;107:djv012 [PubMed]
- Kelloff GJ, Sigman CC. Cancer biomarkers: selecting the right drug for the right patient. Nat Rev Drug Discov 2012;11:201-14. [PubMed]
- Simon R. Clinical trials for predictive medicine: new challenges and paradigms. Clin Trials 2010;7:516-24. [PubMed]
- Parkinson DR, McCormack RT, Keating SM, et al. Evidence of clinical utility: an unmet need in molecular diagnostics for patients with cancer. Clin Cancer Res 2014;20:1428-44. [PubMed]
- Sawyers CL, van't Veer LJ. Reliable and effective diagnostics are keys to accelerating personalized cancer medicine and transforming cancer care: a policy statement from the american association for cancer research. Clin Cancer Res 2014;20:4978-81. [PubMed]
- Poste G. Bring on the biomarkers. Nature 2011;469:156-7. [PubMed]
- Ransohoff DF, Gourlay ML. Sources of bias in specimens for research about molecular markers for cancer. J Clin Oncol 2010;28:698-704. [PubMed]
- Altman DG, McShane LM, Sauerbrei W, et al. Reporting recommendations for tumor marker prognostic studies (REMARK): explanation and elaboration. BMC Med 2012;10:51. [PubMed]
- von Elm E, Altman DG, Egger M, et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Ann Intern Med 2007;147:573-7. [PubMed]
- Bossuyt PM, Reitsma JB, Bruns DE, et al. Towards complete and accurate reporting of studies of diagnostic accuracy: the STARD initiative. BMJ 2003;326:41-4. [PubMed]
- Hoshida Y, Villanueva A, Kobayashi M, et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med 2008;359:1995-2004. [PubMed]
- Hoshida Y, Villanueva A, Sangiovanni A, et al. Prognostic gene expression signature for patients with hepatitis C-related early-stage cirrhosis. Gastroenterology 2013;144:1024-30. [PubMed]
- King LY, Canasto-Chibuque C, Johnson KB, et al. A genomic and clinical prognostic index for hepatitis C-related early-stage cirrhosis that predicts clinical deterioration. Gut 2014; [Epub ahead of print]. [PubMed]
- Fuchs BC, Hoshida Y, Fujii T, et al. Epidermal growth factor receptor inhibition attenuates liver fibrosis and development of hepatocellular carcinoma. Hepatology 2014;59:1577-90. [PubMed]
- Taube SE, Clark GM, Dancey JE, et al. A perspective on challenges and issues in biomarker development and drug and biomarker codevelopment. J Natl Cancer Inst 2009;101:1453-63. [PubMed]
- Watson RW, Kay EW, Smith D. Integrating biobanks: addressing the practical and ethical issues to deliver a valuable tool for cancer research. Nat Rev Cancer 2010;10:646-51. [PubMed]
- National Cancer Institute. NCI Best Practices for Biospecimen Resources. Available online: http://biospecimens.cancer.gov/bestpractices/2011-NCIbestpractices.pdf
- Massett HA, Atkinson NL, Weber D, et al. Assessing the need for a standardized cancer HUman Biobank (caHUB): findings from a national survey with cancer researchers. J Natl Cancer Inst Monogr 2011;2011:8-15.
- Baggerly KA, Morris JS, Coombes KR. Reproducibility of SELDI-TOF protein patterns in serum: comparing datasets from different experiments. Bioinformatics 2004;20:777-85. [PubMed]
- Petricoin EF, Ardekani AM, Hitt BA, et al. Use of proteomic patterns in serum to identify ovarian cancer. Lancet 2002;359:572-7. [PubMed]
- Hernández B, Parnell A, Pennington SR. Why have so few proteomic biomarkers "survived" validation? (Sample size and independent validation considerations). Proteomics 2014;14:1587-92. [PubMed]
- Dupuy A, Simon RM. Critical review of published microarray studies for cancer outcome and guidelines on statistical analysis and reporting. J Natl Cancer Inst 2007;99:147-57. [PubMed]
- Mesirov JP. Computer science. Accessible reproducible research. Science 2010;327:415-6. [PubMed]
- Barrett T, Troup DB, Wilhite SE, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res 2009;37:D885-90. [PubMed]
- Sequist LV, Heist RS, Shaw AT, et al. Implementing multiplexed genotyping of non-small-cell lung cancers into routine clinical practice. Ann Oncol 2011;22:2616-24. [PubMed]
- Zhang Z, Chan DW. The road from discovery to clinical diagnostics: lessons learned from the first FDA-cleared in vitro diagnostic multivariate index assay of proteomic biomarkers. Cancer Epidemiol Biomarkers Prev 2010;19:2995-9. [PubMed]
- Koscielny S. Why most gene expression signatures of tumors have not been useful in the clinic. Sci Transl Med 2010;2:14ps2.
- van de Vijver MJ, He YD, van't Veer LJ, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 2002;347:1999-2009. [PubMed]
- Chia SK, Bramwell VH, Tu D, et al. A 50-gene intrinsic subtype classifier for prognosis and prediction of benefit from adjuvant tamoxifen. Clin Cancer Res 2012;18:4465-72. [PubMed]
- Geiss GK, Bumgarner RE, Birditt B, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 2008;26:317-25. [PubMed]
- FDA. Laboratory Developed Tests. Available online: http://www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/InVitroDiagnostics/ucm407296.htm
- Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 2014;311:1998-2006. [PubMed]
- Hegde M, Bale S, Bayrak-Toydemir P, et al. Reporting incidental findings in genomic scale clinical sequencing--a clinical laboratory perspective: a report of the Association for Molecular Pathology. J Mol Diagn 2015;17:107-17. [PubMed]
- Mardis ER. The $1,000 genome, the $100,000 analysis? Genome Med 2010;2:84. [PubMed]
- April C, Klotzle B, Royce T, et al. Whole-genome gene expression profiling of formalin-fixed, paraffin-embedded tissue samples. PLoS One 2009;4:e8162 [PubMed]
- Reis PP, Waldron L, Goswami RS, et al. mRNA transcript quantification in archival samples using multiplexed, color-coded probes. BMC Biotechnol 2011;11:46. [PubMed]
- Kojima K, April C, Canasto-Chibuque C, et al. Transcriptome profiling of archived sectioned formalin-fixed paraffin-embedded (AS-FFPE) tissue for disease classification. PLoS One 2014;9:e86961 [PubMed]
- Crowley E, Di Nicolantonio F, Loupakis F, et al. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol 2013;10:472-84. [PubMed]
- Plaks V, Koopman CD, Werb Z. Cancer. Circulating tumor cells. Science 2013;341:1186-8. [PubMed]
- Thierry AR, Mouliere F, El Messaoudi S, et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med 2014;20:430-5. [PubMed]
- Smerage JB, Barlow WE, Hortobagyi GN, et al. Circulating tumor cells and response to chemotherapy in metastatic breast cancer: SWOG S0500. J Clin Oncol 2014;32:3483-9. [PubMed]
- Guo QJ, Miils JN, Mason N, et al. Abstract A47: MicroRNA-510 as a predictive marker for response to platinum-based chemotherapy in triple negative breast cancer. Clin Cancer Res 2015;A47.
- Simmer F, Dijkstra J, Venderbosch S, et al. Abstract 534: MicroRNA-143 is a putative predictive biomarker for 5-FU-based chemotherapy in patients with metastatic colorectal cancer. Cancer Res 2014;74:534.
- Teutsch SM, Bradley LA, Palomaki GE, et al. The Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Initiative: methods of the EGAPP Working Group. Genet Med 2009;11:3-14. [PubMed]
- Dowsett M, Cuzick J, Wale C, et al. Prediction of risk of distant recurrence using the 21-gene recurrence score in node-negative and node-positive postmenopausal patients with breast cancer treated with anastrozole or tamoxifen: a TransATAC study. J Clin Oncol 2010;28:1829-34. [PubMed]
- Febbo PG, Ladanyi M, Aldape KD, et al. NCCN Task Force report: Evaluating the clinical utility of tumor markers in oncology. J Natl Compr Canc Netw 2011;9:S1-32; quiz S33.
- . Home-brew tests need regulation. Nature 2014;512:5. [PubMed]
- Rubin EH, Allen JD, Nowak JA, et al. Developing precision medicine in a global world. Clin Cancer Res 2014;20:1419-27. [PubMed]
- National Comprehensive Cancer Network. About the NCCN Biomarkers Compendium®. Available online: http://www.nccn.org/professionals/biomarkers/default.aspx
- Birkeland ML, McClure JS. Optimizing the Clinical Utility of Biomarkers in Oncology: The NCCN Biomarkers Compendium. Arch Pathol Lab Med 2015;139:608-11. [PubMed]
- Wolff AC, Hammond ME, Hicks DG, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol 2013;31:3997-4013. [PubMed]
- FDA. List of Cleared or Approved Companion Diagnostic Devices (In Vitro and Imaging Tools). Available online: http://www.fda.gov/MedicalDevices/ProductsandMedicalProcedures/InVitroDiagnostics/ucm301431.htm
- Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 2010;376:687-97. [PubMed]
- Jordan VC. Selective estrogen receptor modulation: concept and consequences in cancer. Cancer Cell 2004;5:207-13. [PubMed]
- Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol 2008;26:1626-34. [PubMed]
- Dematteo RP, Ballman KV, Antonescu CR, et al. Adjuvant imatinib mesylate after resection of localised, primary gastrointestinal stromal tumour: a randomised, double-blind, placebo-controlled trial. Lancet 2009;373:1097-104. [PubMed]
- Kantarjian H, Sawyers C, Hochhaus A, et al. Hematologic and cytogenetic responses to imatinib mesylate in chronic myelogenous leukemia. N Engl J Med 2002;346:645-52. [PubMed]
- Tallman MS, Andersen JW, Schiffer CA, et al. All-trans-retinoic acid in acute promyelocytic leukemia. N Engl J Med 1997;337:1021-8. [PubMed]
- Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol 2011;12:175-80. [PubMed]
- Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med 2013;368:2385-94. [PubMed]
- Chapman PB, Hauschild A, Robert C, et al. I Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 2011;364:2507-16. [PubMed]
- Mamounas EP, Tang G, Fisher B, et al. Association between the 21-gene recurrence score assay and risk of locoregional recurrence in node-negative, estrogen receptor-positive breast cancer: results from NSABP B-14 and NSABP B-20. J Clin Oncol 2010;28:1677-83. [PubMed]
- Nguyen HG, Welty CJ, Cooperberg MR. Diagnostic associations of gene expression signatures in prostate cancer tissue. Curr Opin Urol 2015;25:65-70. [PubMed]
- You YN, Rustin RB, Sullivan JD. Oncotype DX® colon cancer assay for prediction of recurrence risk in patients with stage II and III colon cancer: A review of the evidence. Surg Oncol 2015;24:61-6. [PubMed]
- Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med 2014;20:548-54. [PubMed]
- Schultz NA, Dehlendorff C, Jensen BV, et al. MicroRNA biomarkers in whole blood for detection of pancreatic cancer. JAMA 2014;311:392-404. [PubMed]
- Rutgers E, Piccart-Gebhart MJ, Bogaerts J, et al. Baseline results of the eortc 10041/mindact trial (microarray in node 0-3 positive disease may avoid chemotherapy). Eur J Cancer 2013;49:S464-5.
- Sharfstein J. FDA regulation of laboratory-developed diagnostic tests: protect the public, advance the science. JAMA 2015;313:667-8. [PubMed]
- Evans JP, Watson MS. Genetic testing and FDA regulation: overregulation threatens the emergence of genomic medicine. JAMA 2015;313:669-70. [PubMed]
- Marx V. Biology: The big challenges of big data. Nature 2013;498:255-60. [PubMed]
- Rehm HL, Berg JS, Brooks LD, et al. Clingen—the clinical genome resource. N Engl J Med 2015;372:2235-42. [PubMed]
- National Humane Genome Research Institute. Electronic Medical Records and Genomics (eMERGE) Network. Available online: http://www.genome.gov/27540473
- Kannry JL, Williams MS. Integration of genomics into the electronic health record: mapping terra incognita. Genet Med 2013;15:757-60. [PubMed]
- Ratner M. Myriad decision aftershocks ripple through biotech. Nat Biotechnol 2013;31:663-5. [PubMed]
- Harrison C. Patenting natural products just got harder. Nat Biotechnol 2014;32:403-4. [PubMed]
- Cole P. Patentability of Genes: A European Union Perspective. Cold Spring Harb Perspect Med 2014;5:a020891 [PubMed]
- Harrison C. Patent watch: Australian court upholds Myriad's gene patent. Nat Rev Drug Discov 2014;13:805. [PubMed]