
Contributions of DNA methylation aberrancies in shaping the cancer epigenome
DNA methylation in mammalian tissues
DNA methylation is an important gene expression regulator, and serves as a guiding force for X chromosome inactivation, cellular differentiation and development, genomic imprinting and the suppression of repetitive elements. In mammalian cells, DNA methylation is mainly restricted to the C-5 position of cytosine in the 5'-CG-3', or CpG, sequence context, and are stable with successive rounds of cell division [reviewed in (1,2)]. DNA methylation in non-CpG regions, CpA, CpC or CpT (generically labeled as CpH), is present in embryonic stem (ES) cells (3). DNA methylation profiles are erased during in embryonic development, and then are re-established as cells develop towards the differentiated, somatic state [reviewed in (4)].
Establishing and maintaining CpG methylation patterns in somatic mammalian cells is accomplished by several DNA methyltransferases (DNMTs) and the co-factor S-adenosylmethionine. DNMT1 is historically considered as a maintenance methyltransferase, as it has an affinity for hemi-methylated DNA and is tightly coordinated to DNA replication machinery [reviewed in (1)]. DNMT3A and DNMT3B were identified as de novo methyltransferases, however, the coordinated efforts of DNMT1, DNMT3A and DNM3B are thought to be required for establishing replicating existing DNA methylation patterns in cancer cells (1). DNMT3A and DNMT3B are believed to selectively anchor nucleosomes containing methylated CpG islands and repetitive elements, suggesting that DNA methylation patterns are influenced by DNA hemimethylation and chromatin cues (5,6). Finally, DNMT3L is only expressed during gametogenesis and embryonic development and serves as a scaffold protein in connecting DNMT3A to nucleosomes (7,8), while DNMT2 functions as a tRNA-methyltransferase (9,10).
Although CpG methylation is an essential regulatory element, it is inherently mutagenic, as 5-methylcytosine (5-mC) undergoes spontaneous deamination to thymine. The rate of 5-mC deamination is approximately an order of magnitude greater than the deamination of unmethylated cytosine to uracil (11-13). As a consequence, CpG content is 20% of what is expected. In addition, approximately 70% of CpG dinucleotides are generally methylated in normal somatic human tissues, representing 4-5% of all cytosines in the human genome, and are generally localized to repetitive elements and regions of low CpG density [reviewed in (12)]. Alternatively, there are regions of the genome, termed CpG islands, which contain their expected number of CpG nucleotides and G:C content. These typically are unmethylated in normal somatic tissues and are frequently located in gene promoter and 5' coding regions (12).
5-hydroxymethylcytosine and DNA demethylation
5-hydroxymethylation (5-hmC) was first described as a product of 5-mC oxidation by TET1 (ten-eleven translocase) [reviewed in (14,15)]. Two additional TET enzymes, TET2 and TET3, were subsequently identified, with each TET enzyme functioning as 2-oxoglutarate- and iron-dependent dioxygenases that are similar in function to several known histone lysine demethylases. TET enzymes can catalyze the conversion of 5-mC to not only 5-hmC, but also the subsequent conversion of 5-hmC to 5-formylcytosine (5-fC) and 5-carboxylcytosine (5-caC). The latter (5-fC and 5-caC) are substrates for thymine DNA glycosylase-mediated base excision repair that results with replacement of the 5-fC and 5-caC base by an unmethylated cytosine. 5-hmC is present at lower levels (<1%) than 5-mC (4-5%), but 5-hmC marks are found at gene promoters, gene bodies and enhancers across tissue types. Specifically, the highest 5-hmC levels are found in brain, colorectal, kidney and liver tissues, whereas 5-hmC levels are substantially lower in heart and breast tissues (16). 5-hmC profiles are also altered in several human cancers [reviewed in (17)], and represent an important step in enzyme-catalyzed DNA demethylation, as well as potential cancer-specific biomarkers (18-22).
Interplay of DNA methylation and chromatin modifications in regulating gene expression
DNA methylation is closely associated with chromatin structure and nucleosome accessibility in regulating gene expression, and as a result, chromatin modifications are also altered in human cancers. Chromatin structure is predominantly described by post-translational modifications of specific amino acids on histone N-terminal tails, in which histone methyltransferases (HMTs), histone acetyltransferases (HATs), histone phosphorylases, and other enzymes catalyze the recognition (readers), addition (writers), removal (erasers) of these functional groups, thereby influencing chromatin structure, and ultimately, gene activity potential [reviewed in (23)]. Chromatin modifications also delineate between genes that display inducible or tissue-specific expression profiles and genes that display constitutive expression and unexpressed genes. Histone lysine monomethylation (H3K4, H3K9, H3K27, H3K79, H4K20) and acetylation (H3K9Ac, H3K14Ac and H3K27Ac) marks correlate with unmethylated DNA and active gene expression. Histone marks associated with gene activation include histone H3 lysine 4 tri-methylation (H3K4me3), H3K36me3 and H3K79me2, while repressed regions of the genome are enriched for H3K9me2, H3K9me3, H3K27me2 and H3K27me3 marks (23-25), which positively correlate with DNA methylation in gene promoters and repetitive elements. Ultimately, the key roles of DNA methylation in regulating chromatin structure are in stabilizing nucleosome position and acting as a repressive mark (1).
DNA methylation alterations in human cancers
DNA methylation alterations are widespread and present in every human cancer type. Specifically, human cancer methylomes generally display global DNA hypomethylation, especially with respect to repetitive elements, low-density CpG regions and lamin-attachment domains (26-34). These DNA hypomethylation events are concomitant with DNA hypermethylation at CpG islands and CpG island shores, which are defined as the regions flanking CpG islands. CpG island shore methylation is also involved in both tissue-specific and cancer-specific differentially methylated regions (35). DNA hypermethylation of promoter/5' CpG islands can correlate with reduction in gene expression, mainly via recruitment of methylated DNA binding proteins (MBDs), together with specific chromatin modifications that result in closed, inactive chromatin marked by H3K27me3 [reviewed in (1,23,36,37)]. Alternatively, gene body DNA methylation correlates with increased gene expression and the presence of H3K36me3 across the gene body region (38-40).
Interestingly, cancer-associated DNA methylation is enriched at genes highlighted for transcriptional repression in embryonic stem (ES) cells via occupancy of the polycomb repressive complex 2 (PRC2), which consists of suppressor of zeste 12 (SUZ12), embryonic ectoderm development (EED) and H3K27me3 marks as a result of enhancer of zeste homolog 2 (EZH2) activity (41,42). These findings suggest a potential stem cell origin of cancer, in which the reversible repression of gene expression in ES cells is ultimately converted into an irreversibly silenced state by DNA hypermethylation and repressive chromatin marks (42). Indeed, tumors are thought to arise from early progenitor, stem-cell like or undifferentiated somatic cells, with activation of pathways related to progenitor/precursor cell growth (43), suggesting that tumors may acquire changes consistent with embryonic cells.
Epigenetic silencing is one mechanism by which genes encoding for tumor suppressors, DNA repair enzymes, and proteins involved in other cellular/regulatory pathways, are inactivated in human cancers. DNA methylation is included with somatic mutations and copy number alterations as means by which individual gene alleles can be inactivated in human cancers. The genome-wide scope of DNA methylation perturbations indicates that these are early events in human carcinogenesis. However, it should be noted that DNA methylation-mediated gene silencing occurs in a small proportion of genes in the cancer methylome. Indeed, less than 10% of hypermethylated genes displayed epigenetic silencing in several reports (44,45). However, this may also be a consequence of tumor cell heterogeneity. Epigenetic silencing may occur in a higher percentage of tumor cells, but may be masked due to both high tumor cell heterogeneity and technological limitations of characterizing single-cell epigenetic silencing events in primary tumors. The advancement of technologies to identify epigenetic silencing events in individual tumor cells will be critical for a complete understanding of the scope of epigenetic silencing as a driver event in human cancers.
In contrast to somatic mutations, DNA methylation alterations are substantially more abundant in human cancers, with approximately 400 hypermethylated genes per cancer genome (46), and therefore represent an abundance of biomarkers for cancer diagnosis, response to treatment and disease monitoring, as well as targets for epigenetic therapies. The latter is an important point, as DNA methylation is reversible through the use of DNA methylation inhibitors. While DNA methylation patterns are heterogeneous within individual loci as well as between individual cells, several gene regions display high frequencies of cancer-associated DNA hypermethylation, therefore allowing for high cancer detection sensitivity and specificity. In addition, while most studies in the field have focused on epigenetic silencing during tumorigenesis, gene body DNA hypermethylation may also be involved in up-regulation of genes in the MYC and metabolic pathways (40).
Candidate epigenetic driver genes and affected signaling pathways
An important aspect of cancer genomics is characterizing specific alterations that drive tumor formation, maintenance and progression. This is especially true with respect to cancer epigenetics, in which DNA methylation aberrancies are widespread in every cancer type. One approach of characterizing potential epigenetic driver genes is to identify genes that are epigenetically silenced in tumor types and subtypes. Epigenetic silencing of key tumor suppressor, regulatory and repair genes has been demonstrated in several cancer types, and as a result, several important cellular signaling pathways are also disrupted. These include DNA repair, RB1/CDK4 cell cycle regulation, WNT/β-catenin, TGF-β, cellular differentiation pathways, as well as others.
CDKN2A (p16INK4A) is frequently silenced via promoter DNA hypermethylation across several human cancers (47,48). The p16 protein binds to cyclin-dependent kinases CDK4 and CDK6, which in turn blocks phosphorylation of retinoblastoma 1 (RB1), thereby allowing the cell to pass through the G1/S cell cycle checkpoint. In this manner, p16 also blocks improper cellular division as a result of DNA damage or oncogenic signaling, however, improper p16 function, whether the result of mutation, deletion or epigenetic silencing, allows the cell to bypass this checkpoint [reviewed in (49)]. For this reason, p16 is considered a tumor suppressor and p16 alterations are thought to be early events in tumorigenesis.
The evolutionarily conserved WNT/β-catenin signaling pathway, commonly referred to as the canonical WNT pathway, is frequently dysregulated in several forms of human cancer. WNT pathway alterations were identified in over 90% of colorectal tumors (50). WNT signaling is predicated upon a multi-protein complex comprised of APC, GSK-3β, axin and β-catenin. In the absence of WNT ligand, binding of WNT ligands to targeted Frizzled receptors leads to activation of the Disheveled protein and GSK-3β inhibition, which stabilizes β-catenin levels [reviewed in (51)]. As a result, β-catenin is localized to the nucleus and consequently binds to transcription factors that in turn induce expression of MYC and CCND1. WNT signaling is inhibited by the absence of ligand, or by the direct binding of WNT antagonists, including WNT inhibitory factors (WIFs) and secreted frizzled-related proteins (SFRPs) to WNT ligands of Frizzled receptors. SFRPs also inhibit cell cycle progression and cellular proliferation. Interestingly, several members of the SFRP gene family, namely SFRP1, SFRP2, SFRP4 and SFRP5, are epigenetically silenced via promoter DNA hypermethylation in several cancer types, most notably in colorectal cancer (52,53).
Mutations in the MSH family of DNA mismatch repair genes are frequent events in human hereditary colorectal cancers (Lynch syndrome), thereby rendering the cancer genome susceptible to mutational burden and microsatellite instability (54). In addition, the mismatch repair gene MLH1 is silenced by promoter DNA hypermethylation in a subset of sporadic colorectal cancers, resulting in the similar impairment of DNA mismatch repair and subsequent expansion of microsatellite repeats (55-57).
BRCA1 and BRCA2 genes are altered in breast and ovarian cancers (58) by several mechanisms, including mutation, copy number and epigenetic silencing as a result of DNA hypermethylation. Sequence alterations in BRCA1 and BRCA2 are evident in over 20% of inherited breast cancer cases, and account for 60-80% lifetime risk of developing breast cancer and 20-40% risk of developing ovarian cancer (59). BRCA1 and BRCA2 mutations account for 50-70% of sporadic breast cancer cases. With these frequencies in mind, BRCA1 and BRCA2 inactivation correlates with genomic instability, chromosomal translocations and pronounced insertions and deletions. BRCA1 is involved in double strand break repair and is involved in guidance of the cell at the G2/S checkpoint. In addition, BRCA1 is involved in several complexes that activate or repress cell cycle arrest, DNA repair and anti-apoptotic processes (59). Inactivation of BRCA1 as a result of mutation, deletion and silencing due to promoter DNA hypermethylation can occur on individual alleles in ovarian and breast cancers (60,61). Indeed, a population-based study of BRCA1 DNA hypermethylation in breast tumors showed a positive correlation of BRCA1 DNA methylation with lower survival rates (62).
Accompanying the concept of a select set of genomic alterations that drive tumorigeneisis is the concept of oncogenic addiction for cancer cell survival. As originally described by Weinstein, oncogenic addiction is thought of as the dependence of the cancer cell on a single oncogenic pathway for survival and the maintenance of the highly proliferative state (63). Interestingly, oncogenic addition supports the notion that targeting these specific addicted pathways can lead to efficacious therapeutic treatments, as these pathways are not active in normal cells. Examples of oncogenes that result in oncogenic addiction are ABL, BRAF, EGFR, HER2, KIT, MET, MYC, RAS, and others, across several forms of human cancer (64). Similarly, an addiction to the absence of tumor suppressor genes also exists in human cancer cells (65). Since tumor suppressor genes are inactivated in many human cancer types, re-activating tumor suppressor genes can also lead to deleterious results for cancer cell survival. Examples of tumor suppressor genes that foster oncogenic addiction when silenced include DLC1, FHIT, PTEN, TP53 and WWOX (65).
The theme of cancer gene addiction can also be applied to DNA methylation, in that DNA methylation of a select set of genes in the cancer genome is absolutely essential for cancer cell growth and survival. In a report De Carvalho and colleagues (66), performed genome-scale DNA methylation analyses of human cancer cells deficient for one or more DNMTs in order to identify those genes that require DNA methylation for cancer cell survival. Indeed, the DNA methylation status of interleukin-1 receptor-associated kinase 3 (IRAK3) is cancer-specific, and correlated with reduced gene expression in cancer cells. Interestingly, IRAK3 inhibits MAPK, NFkB and STAT3 signaling pathways, all of which are activated in several cancer types. IRAK3 promoter undergoes cancer-specific DNA hypermethylation and reduced gene expression in primary tissues across multiple cancer types. Therefore, IRAK3 is an example of an epigenetic driver gene whose DNA methylation is essential for cancer cell survival.
Technological advancements of DNA methylation characterization
Restriction enzyme and PCR-based assays
The discovery of novel targets of cancer-specific DNA methylation and subsequent genome-wide characterization of human cancer methylomes is directly related to technological advancements in measuring DNA methylation changes [reviewed in (12,67)]. Initially, DNA methylation levels were measured globally using high-performance liquid chromatography (HPLC), which can separate cytosine from 5-mC nucleosides. For this reason, 5-mC has been labeled as the 5th base. However, HPLC only can quantitate global levels, but not at specific regions of the genome. The use of methylation-sensitive and methylation-insensitive DNA restriction enzyme isoschizomers (HpaII versus MspI) with PCR in assays such as combined bisulfite restriction analysis (COBRA), HpaII tiny fragment enriched by ligation-mediated PCR (HELP) and methylated CpG island amplification (MCA) provided the ability to study DNA methylation changes at individual CpG sites, however, the surveyed CpG dinucleotides were limited to those that located are at the restriction sites of such enzymes (68-70).
Exploratory screening methods of biomarker discovery include methylation-sensitive arbitrarily primed PCR (MS-AP-PCR), amplification of intermethylated sites (AIMS) and restriction landmark genomic sequencing (RLGS), all of which utilize methylation-sensitive and methylation-insensitive restriction enzyme isoschizomer digestion (71-73). In MS-AP-PCR, genomic DNA digestion is followed by PCR with random CpG-rich primers and gel electrophoresis to identify aberrantly methylated regions that are then characterized by DNA sequencing. The AIMS assay is similar with respect to restriction enzyme treatment, but the methylated DNA ends are then ligated to adapters, and finally amplified by PCR using primers towards the adapter sequences. Like AP-PCR, AIMS PCR products are fractionated by gel electrophoresis to identify differentially methylated fragments that are subsequently sequenced. In RLGS, the digested DNA regions are fractionated on two-dimensional gels to isolate methylated and unmethylated loci. While these assays identify DNA hyper- and hypomethylated loci as biomarkers of disease, they may not be specifically targeted to promoter regions, and therefore may not be efficient in identifying regions that correlate with expression changes.
Since 5-mC marks are lost with PCR amplification of genomic DNA, DNA methylation assays were slower to develop as compared to assays to identify DNA sequence alterations. However, chemically treating genomic DNA with bisulfite results in the conversion of unmethylated cytosines to uracil (and thymine during PCR), while methylated cytosines are unaffected. Therefore, DNA methylation can be interpreted via cytosine versus thymine sequence differences (12). Bisulfite-mediated techniques were subsequently developed, such as candidate gene bisulfite sequencing, methylation-specific PCR (MSP), quantitative MSP (qMSP), MethyLight, pyrosequencing (PSQ) and methylation-sensitive single nucleotide primer extension (MS-SNuPE), in order to rapidly interrogate the DNA methylation status of candidate gene regions in large numbers of primary cancer tissues and cell lines (74-78).
MSP and MethyLight assays provide quick and efficient means for individual research laboratories to measure DNA methylation of virtually any candidate gene region. Gel-based MSP is largely a qualitative measure of DNA methylation, while qMSP and MethyLight are quantitative assays due to the inclusion of SYBR green and non-extending TaqMan fluorescent probes, respectively. In both instances, MSP and MethyLight technologies interrogate regions of concordant DNA methylation across 100-500 base pair PCR amplicons (74,77).
Microarray and next-generation sequencing applications
One caveat of candidate-gene technologies is the requirement of knowing the specific gene regions to interrogate. The union of the release of the human genome reference sequence with the development of microarray and next-generation sequencing technologies has provided the ability for rapid identification of aberrantly methylated candidate gene regions in human cancers. In addition, the combination of methylation-sensitive restriction enzymes, PCR and array hybridization in assays such as methylated CpG island amplification and microarray (MCAM) and differential methylation hybridization (DMH) provide genome-scale analyses of aberrantly methylated DNA regions in cancer tissues and cell lines (79,80).
Another approach for performing genome-scale methylome profiling utilizes enrichment in 5-mC content using antibodies directed towards methylated DNA (MeDIP) and methylated MBDs followed by hybridization to high-density DNA sequence arrays in order to identify hypermethylated genomic DNA regions. These include MeDIP-chip, MBD-chip, the methylated-CpG island recovery assay (MIRA) and comprehensive high-throughput arrays for relative methylation (CHARM) (81-84). This approach has identified cancer-specific DNA methylation events, however, it is not applicable to large numbers of samples and may not accurately identify methylated DNA regions as compared to bisulfite-based methods.
Three Illumina DNA methylation BeadArray platforms, GoldenGate, Infinium HumanMethylation27 (HM27) and Infinium HumanMethylation450 (HM450), provide genome-scale DNA methylation detection of 1,536, 27,578 and 482,421 CpG dinucleotides, respectively, with the ability of surveying DNA methylation levels of large numbers of samples (85-89). Currently, the Illumina HM450 BeadArray is the only commercially available DNA methylation array platform, and is regarded as a cost-effective, high-throughput method for biomarker and tumor subtype identification. The HM450 platform can also be used in conjunction with nucleosome positioning (accessible) and oxidative bisulfite based 5-hmC profiling assays to obtain integrated views of the cancer epigenome (90-93).
Next-generation based sequencing approaches (-seq), including MBD-seq, MeDIP-seq, whole-genome bisulfite sequencing (WGBS) and reduced representation bisulfite sequencing (RRBS), have become more cost-effective and therefore increasingly utilized to obtain more comprehensive DNA methylation maps. Using WGBS, DNA methylation information can be obtained for nearly all of the 28 million total CpG sites in the human genome (26,32,33). However, WGBS requires substantial sequencing depth (4-30× genome coverage) to obtain high-quality and interpretative data, and is challenging with respect to mapping CpG-rich sequences and repetitive element regions of the genome. Alternatively, RRBS involves the use of methylation-specific restriction enzyme isoschizomers (HpaII versus MspI), followed by library construction, for profiling of approximately 1% of the human genome, thereby reducing the overall required sequencing while determining the DNA methylation status of 1-2 million CpG dinucleotides, mostly in CpG islands and gene promoters (94,95). Finally, the recently reported NOMe-seq (nucleosome occupancy and methylation) technology displays the ability to concurrently determine both DNA methylation and nucleosome occupancy (96).
Sensitive detection of human cancers using DNA methylation-based approaches
Since cancer-specific DNA methylation alterations are stable and present in all forms of human cancer, DNA methylation biomarkers have a promising utility for cancer diagnostics, disease monitoring, treatment response, as well as prediction of disease risk and survival. With these applications in mind, there is tremendous interest in identifying cancer-specific events for early detection purposes, as cancer-derived DNA is present in the bloodstream of cancer patients. Additional bodily fluids, including urine sediment, sputum and fecal matter, also represent promising media for capturing and quantifying cancer-specific genomic alterations.
Colorectal cancer detection using the SEPT9 DNA methylation marker in cell-free DNA isolated from plasma has shown great promise as an early detection biomarker. SEPT9 DNA methylation was identified after a screening cancer-specific DNA hypermethylation using MS-AP-PCR and MCA approaches (97). The SEPT9 assay displays a mean 75% sensitivity and 87% specificity after testing of thousands of samples using a variety of protocols in the United States and Europe [summarized in (98)]. Specifically, the second iteration of the assay displayed mean detection sensitivities of 67%, 83%, 84% and 100% for stage I, II, III and IV colorectal cancers, respectively (99). Moreover, the assay is positive in nearly 30% of advanced adenomas, with 58% sensitivity and 82% specificity (99). Overall, the SEPT9 DNA methylation assay is currently distributed as commercially available diagnostic kits under the names Epi proColon (Epigenomics), mS9 (Abbott) and ColoVantage (Quest) (100,101).
Cancer-derived, methylated DNA detection in urine sediment is also a promising detection method for bladder and prostate cancers. Due to the large volume of urine available and the ease of sample collection and DNA isolation, urine is an attractive media for disease detection, surveillance, recurrence and response to treatment. Indeed, Su et al. recently described a panel of three markers comprised of SOX1 and IRAK3 DNA hypermethylation and LINE-1 repetitive element DNA hypomethylation that predict tumor recurrence superior to cytology and cystoscopy, which are regarded as the gold standard for bladder cancer surveillance and monitoring (102). Several DNA methylation-based biomarkers of prostate cancer have been identified in urine, including APC, GSTP1, HOXD3 and TGBR2 (103,104), while detection of RASSF1 DNA hypermethylation in urine correlated with prostate tumor recurrence (105).
Digital PCR methods have also improved the discovery of cancer-associated DNA methylation alterations. Bisulfite-based PCR assays performed in individual assay wells are largely limited in detection sensitivity and resolution. In contrast, digital PCR-based assays, in which DNA molecules are PCR-amplified over multiple reaction wells, allow for an accounting of individual methylated DNA molecules (106). This is accomplished since template and non-template molecules are sequestered into individual reaction wells, thereby reducing PCR inhibition and improving the signal to noise ratio.
One application of digital PCR to DNA methylation technology is Digital MethyLight, which results in improved detection sensitivity and quantitative accuracy of individual methylated template DNA molecules (107). Digital MethyLight is not only compatible with conventional real-time PCR platforms, but also with microfluidic and digital droplet PCR platforms (107,108). Digital MethyLight was first utilized to quantitate individual cancer-derived DNA hypermethylation events in CLDN5, FOXE1 and RUNX3 in serum of breast cancer patients (107). Moreover, Campan and colleagues identified DNA methylation of IFFO1 in ovarian cancer patients after Illumina Infinium DNA methylation screening of ovarian tumors and non-tumor tissues (109). Interestingly, tumor-derived IFFO1 detection in patient serum correlated with CA-125 levels currently used for determining disease burden and relapse after tumor resection (109). Similarly, Illumina Infinium DNA methylation screening was used to identify THBD and C9orf50 DNA methylation in colorectal tumors, as well as their potential utility as early-detection markers of colorectal cancer in plasma and serum (110). THBD and C9orf50 DNA methylation outperformed carcinoembryonic antigen (CEA), a clinically-approved marker for colorectal cancer detection in blood (111,112), in terms of tumor detection sensitivities (110).
DNA methylation characterization of human cancers from multi-institutional consortia
As a result of technological advancements in surveying human cancer genomes, molecular profiling of large numbers of primary tumors in order to paint integrative molecular portraits of individual cancer types, is currently attainable. These integrated views, coupled with patient clinical information and large sample numbers, can be utilized for identifying novel molecular-based tumor subgroups and therapeutic approaches. However, determining these features in a genome-wide manner is challenging for several reasons, including the costs of generating and analyzing molecular data, the requirement for stratifying molecular data for large numbers of samples as functions of age, gender, race/ethnicity, environmental exposures and family history. Moreover, genomic features of human cancers also require tissue-specific interrogations, thereby increasing experimental complexity and scope. Substantial laboratory and bioinformatics expertise is also essential, and therefore, is challenging to complete in isolation. Genomic, epigenomic and transcriptomic alterations do not exist in isolation, since the cancer genome typically displays an abundance of structural and regulatory aberrations, and therefore, integrative methods and approaches are required to obtain a complete picture of the cancer genome. The cancer genome is not only confounded by the challenges of identifying linkages between DNA sequence, epigenetics and expression data, but also the issues of genetic/chromosomal instability, tumor cell heterogeneity, multilevel selection and the complex nature of cancer evolution (113-115).
In recognizing these challenges, several multi-institutional consortia have been organized for efficient molecular characterization and analyses. These include Encyclopedia of DNA Elements (ENCODE) (116-118), International Cancer Genomics Consortium, International Human Epigenome Consortium (119), NIH Roadmap Epigenomics Initiative (120-122) and The Cancer Genome Atlas (TCGA) have been vital in identifying genomic and epigenomic alterations in normal tissues, cancer tissues and cancer cell lines.
In particular, TCGA has been prolific in generating and analyzing genomic, epigenomic and transcriptomic data for over 11,000 primary tumors and 1,000 normal-adjacent tissues spanning 30 human cancer types (Table 1) (137). In order to detect low-penetrating genomic alterations, TCGA accrued 500 tumors for most tumor types. Molecular profiling includes mutation detection (whole-genome and whole-exome sequencing), gene expression profiling (RNA-seq and miRNA-seq), somatic copy number alterations (single nucleotide polymorphism arrays), and DNA methylation profiling (Illumina BeadArrays) (85-88). The unique and powerful aspects of the TCGA collections stem from large sample numbers, centralized pathological review, selection of samples with a high fraction of tumor nuclei, genomic characterizations over multiple assay platforms using nucleic acids isolated from the same tissue samples and data integration for high-level pathway interpretations. TCGA has published high profile, integrative analyses of 16 individual tumor types (45,50,60,61,123-135), and is currently pursuing similar analyses for the remaining tumor types, as well integrated analyses of molecular data across multiple cancer types (136,138).
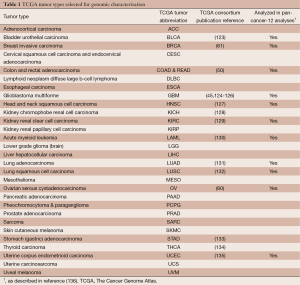
Full table
CpG island methylator phenotypes (CIMPs) as unique and clinically relevant tumor subgroups
Currently, there is wide interest in identifying and characterizing novel DNA methylation-based subgroups of tumors with the goal of determining their relationships to clinical features and other genomic alterations for diagnostic and treatment purposes. In 1999, Toyota and colleagues first described a distinct subset of human colorectal cancers with extensive DNA hypermethylation of a subset of CpG islands that remained unmethylated in the remaining colorectal tumors (139), and are therefore unique from general cancer-specific DNA methylation for a specific tumor type. These tumors, representing approximately 15% of all colorectal cancers, are classified as positive for a CIMP. Colorectal CIMP-positive tumors, currently referred to as CIMP-high (CIMP-H), are enriched for BRAFV600E mutations, microsatellite instability, MLH1 DNA hypermethylation, a hypermutated profile and the absence of copy number alterations and KRAS mutations. Colorectal CIMPs are largely located in the proximal colon, are more prevalent in patients of older age and female gender, family history and demonstrate improved survival (44,50,140-143). In addition, CIMP status in female patients positively correlated with increased pack years of smoking and obesity (144).
In contrast to CIMP-H tumors, an additional CIMP-like subgroup, described as CIMP-low (CIMP-L) and CIMP2, has been identified in a subset of colorectal tumors (50,145,146). CIMP-L tumors display attenuated CIMP-associated DNA methylation, mixed microsatellite stability, extensive copy number alterations and enrichment in KRAS mutations (44,50,143,145). TCGA confirmed the CIMP-H and CIMP-L subgroups, along with their associations to the previously described molecular features (50).
The TCGA consortium was instrumental in characterizing a novel CIMP subgroup (G-CIMP) in glioblastoma (grade IV glioma), which is present in approximately 15% of GBM cases, and is nearly completely correlated with a specific heterogeneous point mutation in the IDH1 gene (IDH1R132H) (45). All TCGA primary GBM tumors with an IDH1 mutation (IDH1MUT) are G-CIMP, however, a small number of G-CIMP tumors are wild type for IDH1 (IDH1WT). G-CIMP tumors are enriched for TP53 alterations, reduced copy number alterations, and correlated with improved survival and younger patient age.
The association of G-CIMP with IDH1 mutations connected two seemingly disparate aspects of cellular biology. While IDH1WT functions in the citric acid cycle by converting isocitrate to alpha-ketoglutarate (α-KG) [reviewed in (147)], IDH1MUT also catalyzes the conversion of α-KG to D-2-hydroxyglutarate (2-HG), and importantly, inhibits TET function and subsequent DNA demethylation. TET inhibition as a result of IDH1MUT supports the hypermethylated DNA landscape in G-CIMP tumors, and this landmark mechanistic discovery has great clinical promise, not only for diagnostic purposes, but also the potential use of epigenetic therapies for treating patients with G-CIMP tumors.
TCGA also recently characterized two gastric cancer CIMP subgroups report (133). The first, gastric CIMP, displays a colorectal-like CIMP DNA methylation profile, together with hypermutation, MSI and MLH1 epigenetic silencing. The second set of gastric CIMP tumors, EBV-CIMP, is associated with EBV infection, is present in nearly 10% of gastric cancer cases, and display extensive CIMP-DNA methylation even beyond that of the Gastric CIMP group. EBV-CIMP is also enriched for mutations of the chromatin remodeler ARID1A, as well as CDKN2A silencing. While the correlation of EBV infection with the EBV-CIMP subgroup is currently unknown, this represents another powerful connection between a novel DNA methylation-based subgroup and unique genomic, clinical and biological features.
CIMP subgroups of breast (B-CIMP) (61) and endometrial (E-CIMP) (135) cancers have also been characterized. The TCGA integrated breast cancer report showed that B-CIMP tumors were positive for estrogen and progesterone receptor expression, as well as for HER2 expression, and were enriched for epigenetic silencing of genes in the Wnt-signaling pathway (61), as also described for colorectal tumors (50). E-CIMP tumors are similar to colorectal CIMP tumors in that both display hypermutation, MLH1 promoter DNA hypermethylation, MSI, and the absence of both TP53 somatic mutations and copy number alterations (50,135). Finally, it should be noted that neither E-CIMP nor B-CIMP tumors harbor BRAFV600E or IDH1 mutations, as described in colorectal and glioma CIMP tumors, respectively, pointing to the hypothesis that individual CIMPs may arise from several possible overlapping and non-overlapping molecular mechanisms.
Cancer-associated DNA methylation alterations as targets for epigenetic-based therapeutics
An important aspect of cancer-associated epigenetic alterations that should not be overlooked is that unlike somatic mutations and copy number alterations, DNA methylation and histone modifications are reversible, and therefore, aberrant epigenetic profiles can be corrected using inhibitors to DNMTs and histone modifiers. The first-generation epigenetic inhibitors based on DNMT inhibition, 5-azacytidine (5-Aza-CR, Vidaza) and 5-aza-2'-deoxycytidine (5-Aza-CdR, decitabine, dacogen), have been approved for treatment of myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML) patients (148,149). In addition, a second-generation inhibitor, zebularine (pyrimidin-2-one β-D-ribofuranoside), also inhibited DNA methylation after oral administration in both in vitro and in vivo settings (150,151).
The treatment of cell line models of human breast, colon and lung tumors with low doses of vidaza resulted in the re-expression of genes specific for immune response and cancer testis antigens, including interferon signaling, antigen processing and cytokines/chemokines (36,152,153), and down regulation of oncogenic signaling pathways, such as MYC and metabolic pathways (40). Moreover, combinatorial treatments of histone deacetylase (HDAC) inhibitors, together with low doses of vidaza over several treatment cycles, also induced gene expression of genes involved in cell cycle regulation, cytoskeletal organization and DNA damage response (154). Indeed, clinical trials in which vidaza and the HDAC inhibitor entinostat were administered to patients with recurrent non-small cell lung cancer (NSCLC) were overall well tolerated, and showed prolonged patient survival (155).
In support of advancing clinical trials with novel epigenetic-based therapies, the recently-introduced DNA methylation inhibitor S110 (AzapG), a dinucleotide consisting of 5-aza-2'-deoxycytidine connected upstream to a deoxyguanosine, was shown to be an effective DNA methylation inhibitor by resisting cytidine deaminase degradation in both in vitro and in vivo settings, and showed improved stability and reduced toxicity compared to aza-substituted mononucleosides (156,157), Currently, S110 is being analyzed for potential clinical utility in phase II clinical trials for treatment of ovarian, liver and colorectal cancers, as well as AML, MDS, either alone, or in combination with other therapeutic agents.
Conclusions
DNA methylation is a complex regulatory element, and it is not surprising that DNA methylation is dysregulated in virtually every tumor type. As a result, gene expression and subsequent signaling pathways are affected, and implicates DNA methylation alterations as early events in tumorigenesis. Importantly, DNA methylation is now looked upon as a major genomic feature of human cancers, together with somatic mutations and copy number changes. Indeed, genes are inactivated by individual types of alterations, as well as by multiple alterations, satisfying Knudsen’s two-hit hypothesis. In addition, DNA methylation aberrancies represent promising cancer therapeutic targets, since DNA methylation profiles are reversible through the use of inhibitors. This feature of cancer epigenomics is important, as DNA methylation inhibitors can be utilized for exploiting addictions to oncogenes and the absence of tumor suppressors. DNA methylation markings can also have clinical utility in their development as cancer biomarkers. DNA methylation-based biomarkers have been described in several reports as indicators of tumor presence, as well as predictors of recurrence, progression. Moreover, DNA methylation biomarkers can be detected in several biological fluids, such as blood, sputum and urine, for early detection of human cancers.
Finally, DNA methylation alterations can now be characterized across the genome in large numbers of primary tissues using next-generation sequencing and microarray methods. Moreover, multi-institutional consortia, including ENCODE, the Epigenetics Roadmap Initiative and TCGA, have been instrumental in generating diverse sets of epigenomic data on large numbers of normal and tumor cell lines and tissues. These publically available datasets have been important in cancer epigenomic profiling and validation efforts, as well as identifying tumor subgroups. In support of this, DNA methylation based subgroups of individual tumor types have been characterized. These are primarily classified as CIMP, and represent subsets of tumors with accentuated cancer-specific DNA methylation profiles. Importantly, CIMPs are clinically relevant, and correlate with patient age, survival and other genomic features.
In summary, DNA methylation represents an important aspect of cancer genomics, based on their roles in gene regulation, biomarkers and therapeutic targets. The improvement of whole-genome sequencing technologies, especially in determining the epigenomic profiles of large, population-based tumor collections, as well as single cells and cell-free tumor-derived DNA in biological fluids, will provide a wealth of information for obtaining higher-resolution maps of the cancer epigenome for refined cancer detection, monitoring, surveillance and targets of therapeutic efficacy.
Acknowledgments
We thank the tireless efforts of thousands of basic and translational scientists and clinicians for advancing the field of cancer epigenetics.
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editor (Jian-Bing Fan) for the series “Application of Genomic Technologies in Cancer Research” published in Translational Cancer Research. The article has undergone external peer review.
Conflicts of Interest: DJ Weisenberger is a consultant for Zymo Research, which distributes commercially available products for DNA methylation-based experiments. Zymo Research did not sponsor this review.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 2012;13:484-92. [PubMed]
- Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science 2001;293:1068-70. [PubMed]
- Ramsahoye BH, Biniszkiewicz D, Lyko F, et al. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci U S A 2000;97:5237-42. [PubMed]
- Morgan HD, Santos F, Green K, et al. Epigenetic reprogramming in mammals. Hum Mol Genet 2005;14:R47-58. [PubMed]
- Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet 2009;10:805-811. [PubMed]
- Sharma S, De Carvalho DD, Jeong S, et al. Nucleosomes containing methylated DNA stabilize DNA methyltransferases 3A/3B and ensure faithful epigenetic inheritance. PLoS Genet 2011;7:e1001286 [PubMed]
- Bourc҆his D, Xu GL, Lin CS, et al. Dnmt3L and the establishment of maternal genomic imprints. Science 2001;294:2536-9.
- Hata K, Okano M, Lei H, et al. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 2002;129:1983-93. [PubMed]
- Goll MG, Kirpekar F, Maggert KA, et al. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 2006;311:395-8. [PubMed]
- Schaefer M, Pollex T, Hanna K, et al. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev 2010;24:1590-5. [PubMed]
- Frederico LA, Kunkel TA, Shaw BR. A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry 1990;29:2532-7. [PubMed]
- Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer 2003;3:253-66. [PubMed]
- Shen JC, Rideout WM 3rd, Jones PA. The rate of hydrolytic deamination of 5-methylcytosine in double-stranded DNA. Nucleic Acids Res 1994;22:972-6. [PubMed]
- Pfeifer GP, Xiong W, Hahn MA, et al. The role of 5-hydroxymethylcytosine in human cancer. Cell Tissue Res 2014;356:631-41. [PubMed]
- Rodger EJ, Chatterjee A, Morison IM. 5-hydroxymethylcytosine: a potential therapeutic target in cancer. Epigenomics 2014;6:503-14. [PubMed]
- Li W, Liu M. Distribution of 5-hydroxymethylcytosine in different human tissues. J Nucleic Acids 2011;2011:870726.
- Ficz G, Gribben JG. Loss of 5-hydroxymethylcytosine in cancer: cause or consequence? Genomics 2014;104:352-7. [PubMed]
- Hahn MA, Qiu R, Wu X, et al. Dynamics of 5-hydroxymethylcytosine and chromatin marks in Mammalian neurogenesis. Cell Rep 2013;3:291-300. [PubMed]
- Pfeifer GP, Kadam S, Jin SG. 5-hydroxymethylcytosine and its potential roles in development and cancer. Epigenetics Chromatin 2013;6:10. [PubMed]
- Haffner MC, Chaux A, Meeker AK, et al. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget 2011;2:627-37. [PubMed]
- Kraus TF, Globisch D, Wagner M, et al. Low values of 5-hydroxymethylcytosine (5hmC), the "sixth base," are associated with anaplasia in human brain tumors. Int J Cancer 2012;131:1577-90. [PubMed]
- Kudo Y, Tateishi K, Yamamoto K, et al. Loss of 5-hydroxymethylcytosine is accompanied with malignant cellular transformation. Cancer Sci 2012;103:670-6. [PubMed]
- Shen H, Laird PW. Interplay between the cancer genome and epigenome. Cell 2013;153:38-55. [PubMed]
- Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell 2007;129:823-37. [PubMed]
- Steger DJ, Lefterova MI, Ying L, et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol 2008;28:2825-39. [PubMed]
- Berman BP, Weisenberger DJ, Aman JF, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet 2012;44:40-6. [PubMed]
- Diala ES, Hoffman RM. Hypomethylation of HeLa cell DNA and the absence of 5-methylcytosine in SV40 and adenovirus (type 2) DNA: analysis by HPLC. Biochem Biophys Res Commun 1982;107:19-26. [PubMed]
- Ehrlich M, Gama-Sosa MA, Huang LH, et al. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res 1982;10:2709-21. [PubMed]
- Ehrlich M, Wang RY. 5-Methylcytosine in eukaryotic DNA. Science 1981;212:1350-7. [PubMed]
- Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 1983;301:89-92. [PubMed]
- Gama-Sosa MA, Slagel VA, Trewyn RW, et al. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res 1983;11:6883-94. [PubMed]
- Hansen KD, Timp W, Bravo HC, et al. Increased methylation variation in epigenetic domains across cancer types. Nat Genet 2011;43:768-75. [PubMed]
- Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009;462:315-22. [PubMed]
- Weisenberger DJ, Campan M, Long TI, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res 2005;33:6823-36. [PubMed]
- Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 2009;41:178-86. [PubMed]
- Baylin SB. The cancer epigenome: its origins, contributions to tumorigenesis, and translational implications. Proc Am Thorac Soc 2012;9:64-5. [PubMed]
- Esteller M, Corn PG, Baylin SB, et al. A gene hypermethylation profile of human cancer. Cancer Res 2001;61:3225-9. [PubMed]
- Bender CM, Gonzalgo ML, Gonzales FA, et al. Roles of cell division and gene transcription in the methylation of CpG islands. Mol Cell Biol 1999;19:6690-8. [PubMed]
- Kulis M, Heath S, Bibikova M, et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet 2012;44:1236-42. [PubMed]
- Yang X, Han H, De Carvalho DD, et al. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell 2014;26:577-90. [PubMed]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature 2011;469:343-9. [PubMed]
- Widschwendter M, Fiegl H, Egle D, et al. Epigenetic stem cell signature in cancer. Nat Genet 2007;39:157-8. [PubMed]
- Jin B, Li Y, Robertson KD. DNA methylation: superior or subordinate in the epigenetic hierarchy? Genes Cancer 2011;2:607-17. [PubMed]
- Hinoue T, Weisenberger DJ, Lange CP, et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res 2012;22:271-82. [PubMed]
- Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 2010;17:510-22. [PubMed]
- Schuebel KE, Chen W, Cope L, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet 2007;3:1709-23. [PubMed]
- Herman JG, Merlo A, Mao L, et al. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res 1995;55:4525-30. [PubMed]
- Merlo A, Herman JG, Mao L, et al. 5’ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med 1995;1:686-92. [PubMed]
- Kamb A. Cell-cycle regulators and cancer. Trends Genet 1995;11:136-40. [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012;487:330-7. [PubMed]
- Serman L, Nikuseva Martic T, Serman A, et al. Epigenetic alterations of the Wnt signaling pathway in cancer: a mini review. Bosn J Basic Med Sci 2014;14:191-4. [PubMed]
- Surana R, Sikka S, Cai W, et al. Secreted frizzled related proteins: Implications in cancers. BiochimBiophys Acta 2014;1845:53-65.
- Suzuki H, Watkins DN, Jair KW, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet 2004;36:417-22. [PubMed]
- Rustgi AK. The genetics of hereditary colon cancer. Genes Dev 2007;21:2525-38. [PubMed]
- Jass JR. HNPCC and sporadic MSI-H colorectal cancer: a review of the morphological similarities and differences. Fam Cancer 2004;3:93-100. [PubMed]
- Jass JR. Sporadic versus hereditary forms of colorectal cancer with the DNA microsatellite instability phenotype: to ‘lump’ or ‘split’? Fam Cancer 2004;3:83. [PubMed]
- Poynter JN, Siegmund KD, Weisenberger DJ, et al. Molecular characterization of MSI-H colorectal cancer by MLHI promoter methylation, immunohistochemistry, and mismatch repair germline mutation screening. Cancer Epidemiol Biomarkers Prev 2008;17:3208-15. [PubMed]
- Hilton JL, Geisler JP, Rathe JA, et al. Inactivation of BRCA1 and BRCA2 in ovarian cancer. J Natl Cancer Inst 2002;94:1396-406. [PubMed]
- Savage KI, Harkin DP. BRCA1, a ‘complex’ protein involved in the maintenance of genomic stability. FEBS J 2015;282:630-46. [PubMed]
- The Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609-15. [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61-70. [PubMed]
- Wu L, Wang F, Xu R, et al. Promoter methylation of BRCA1 in the prognosis of breast cancer: a meta-analysis. Breast Cancer Res Treat 2013;142:619-27. [PubMed]
- Weinstein IB. Disorders in cell circuitry during multistage carcinogenesis: the role of homeostasis. Carcinogenesis 2000;21:857-64. [PubMed]
- Sharma SV, Settleman J. Oncogene addiction: setting the stage for molecularly targeted cancer therapy. Genes Dev 2007;21:3214-31. [PubMed]
- Weinstein IB. Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science 2002;297:63-4. [PubMed]
- De Carvalho DD, Sharma S, You JS, et al. DNA methylation screening identifies driver epigenetic events of cancer cell survival. Cancer Cell 2012;21:655-67. [PubMed]
- Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet 2010;11:191-203. [PubMed]
- Khulan B, Thompson RF, Ye K, et al. Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome Res 2006;16:1046-55. [PubMed]
- Toyota M, Ho C, Ahuja N, et al. Identification of differentially methylated sequences in colorectal cancer by methylated CpG island amplification. Cancer Res 1999;59:2307-12. [PubMed]
- Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res 1997;25:2532-4. [PubMed]
- Costello JF, Hong C, Plass C, et al. Restriction landmark genomic scanning: analysis of CpG islands in genomes by 2D gel electrophoresis. Methods Mol Biol 2009;507:131-48. [PubMed]
- Frigola J, Ribas M, Risques RA, et al. Methylome profiling of cancer cells by amplification of inter-methylated sites (AIMS). Nucleic Acids Res 2002;30:e28 [PubMed]
- Gonzalgo ML, Liang G, Spruck CH 3rd, et al. Identification and characterization of differentially methylated regions of genomic DNA by methylation-sensitive arbitrarily primed PCR. Cancer Res 1997;57:594-9. [PubMed]
- Eads CA, Danenberg KD, Kawakami K, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res 2000;28:E32 [PubMed]
- Gonzalgo ML, Jones PA. Rapid quantitation of methylation differences at specific sites using methylation-sensitive single nucleotide primer extension (Ms-SNuPE). Nucleic Acids Res 1997;25:2529-31. [PubMed]
- Harden SV, Sanderson H, Goodman SN, et al. Quantitative GSTP1 methylation and the detection of prostate adenocarcinoma in sextant biopsies. J Natl Cancer Inst 2003;95:1634-7. [PubMed]
- Herman JG, Graff JR, Myohanen S, et al. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A 1996;93:9821-6. [PubMed]
- Yang AS, Estecio MR, Doshi K, et al. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res 2004;32:e38 [PubMed]
- Estécio MR, Yan PS, Ibrahim AE, et al. High-throughput methylation profiling by MCA coupled to CpG island microarray. Genome Res 2007;17:1529-36. [PubMed]
- Huang TH, Perry MR, Laux DE. Methylation profiling of CpG islands in human breast cancer cells. Hum Mol Genet 1999;8:459-70. [PubMed]
- Irizarry RA, Ladd-Acosta C, Carvalho B, et al. Comprehensive high-throughput arrays for relative methylation (CHARM). Genome Res 2008;18:780-90. [PubMed]
- Jacinto FV, Ballestar E, Esteller M. Methyl-DNA immunoprecipitation (MeDIP): hunting down the DNA methylome. BioTechniques 2008;44:35-43. [PubMed]
- Zilberman D, Henikoff S. Genome-wide analysis of DNA methylation patterns. Development 2007;134:3959-65. [PubMed]
- Rauch T, Pfeifer GP. Methylated-CpG island recovery assay: a new technique for the rapid detection of methylated-CpG islands in cancer. Lab Invest 2005;85:1172-80. [PubMed]
- Bibikova M, Barnes B, Tsan C, et al. High density DNA methylation array with single CpG site resolution. Genomics 2011;98:288-95. [PubMed]
- Bibikova M, Fan JB. GoldenGate assay for DNA methylation profiling. Methods Mol Biol. 2009;507:149-163. [PubMed]
- Bibikova M, Fan JB. Genome-wide DNA methylation profiling. Wiley Interdiscip Rev Syst Biol Med 2010;2:210-23. [PubMed]
- Bibikova M, Le J, Barnes B, et al. Genome-wide DNA methylation profiling using Infinium(R) assay. Epigenomics 2009;1:177-200. [PubMed]
- Bibikova M, Lin Z, Zhou L, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res 2006;16:383-93. [PubMed]
- Field SF, Beraldi D, Bachman M, et al. Accurate Measurement of 5-Methylcytosine and 5-Hydroxymethylcytosine in Human Cerebellum DNA by Oxidative Bisulfite on an Array (OxBS-Array). PloS One 2015;10:e0118202 [PubMed]
- Nazor KL, Boland MJ, Bibikova M, et al. Application of a low cost array-based technique - TAB-Array - for quantifying and mapping both 5mC and 5hmC at single base resolution in human pluripotent stem cells. Genomics 2014;104:358-67. [PubMed]
- Stewart SK, Morris TJ, Guilhamon P, et al. oxBS-450K: a method for analysing hydroxymethylation using 450K BeadChips. Methods 2015;72:9-15. [PubMed]
- Yuan T, Jiao Y, de Jong S, et al. An integrative multi-scale analysis of the dynamic DNA methylation landscape in aging. PLoS Genet 2015;11:e1004996 [PubMed]
- Meissner A, Gnirke A, Bell GW, et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res 2005;33:5868-77. [PubMed]
- Gu H, Bock C, Mikkelsen TS, et al. Genome-scale DNA methylation mapping of clinical samples at single-nucleotide resolution. Nat Methods 2010;7:133-6. [PubMed]
- Kelly TK, Liu Y, Lay FD, et al. Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res 2012;22:2497-506. [PubMed]
- Lofton-Day C, Model F, Devos T, et al. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem 2008;54:414-23. [PubMed]
- Li Y, Song L, Gong Y, et al. Detection of colorectal cancer by DNA methylation biomarker SEPT9: past, present and future. Biomark Med 2014;8:755-69. [PubMed]
- Jin P, Kang Q, Wang X, et al. Performance of a second generation methylated SEPT9 test in detecting colorectal neoplasm. J Gastroenterol Hepatol 2015;30:830-3. [PubMed]
- Payne SR. From discovery to the clinic: the novel DNA methylation biomarker SEPT9 for the detection of colorectal cancer in blood. Epigenomics 2010;2:575-85. [PubMed]
- Heichman KA. Blood-based testing for colorectal cancer screening. Mol Diagn Ther 2014;18:127-35. [PubMed]
- Su SF, de Castro Abreu AL, Chihara Y, et al. A panel of three markers hyper- and hypomethylated in urine sediments accurately predicts bladder cancer recurrence. Clin Cancer Res 2014;20:1978-89. [PubMed]
- Jatkoe TA, Karnes RJ, Freedland SJ, et al. A urine-based methylation signature for risk stratification within low-risk prostate cancer. Br J Cancer 2015;112:802-8. [PubMed]
- Olkhov-Mitsel E, Zdravic D, Kron K, et al. Novel multiplex MethyLight protocol for detection of DNA methylation in patient tissues and bodily fluids. Sci Rep 2014;4:4432. [PubMed]
- Daniunaite K, Jarmalaite S, Kalinauskaite N, et al. Prognostic value of RASSF1 promoter methylation in prostate cancer. J Urol 2014;192:1849-55. [PubMed]
- Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci U S A 1999;96:9236-41. [PubMed]
- Weisenberger DJ, Trinh BN, Campan M, et al. DNA methylation analysis by digital bisulfite genomic sequencing and digital MethyLight. Nucleic Acids Res 2008;36:4689-98. [PubMed]
- Wiencke JK, Bracci PM, Hsuang G, et al. A comparison of DNA methylation specific droplet digital PCR (ddPCR) and real time qPCR with flow cytometry in characterizing human T cells in peripheral blood. Epigenetics 2014;9:1360-5. [PubMed]
- Campan M, Moffitt M, Houshdaran S, et al. Genome-scale screen for DNA methylation-based detection markers for ovarian cancer. PloS One 2011;6:e28141 [PubMed]
- Lange CP, Campan M, Hinoue T, Slingerland H, et al. Genome-scale discovery of DNA-methylation biomarkers for blood-based detection of colorectal cancer. PloS One 2012;7:e50266 [PubMed]
- Goslin R, O’Brien MJ, Steele G, et al. Correlation of Plasma CEA and CEA tissue staining in poorly differentiated colorectal cancer. Am J Med 1981;71:246-53. [PubMed]
- O’Brien MJ, Zamcheck N, Burke B, et al. Immunocytochemical localization of carcinoembryonic antigen in benign and malignant colorectal tissues. Assessment of diagnostic value. Am J Clin Pathol 1981;75:283-90. [PubMed]
- Heng HH, Bremer SW, Stevens JB, et al. Chromosomal instability (CIN): what it is and why it is crucial to cancer evolution. Cancer Metastasis Rev 2013;32:325-40. [PubMed]
- Heng HH, Bremer SW, Stevens JB, et al. Genetic and epigenetic heterogeneity in cancer: a genome-centric perspective. J Cell Physiol 2009;220:538-47. [PubMed]
- Huang S. Genetic and non-genetic instability in tumor progression: link between the fitness landscape and the epigenetic landscape of cancer cells. Cancer Metastasis Rev 2013;32:423-48. [PubMed]
- ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 2004;306:636-40. [PubMed]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57-74. [PubMed]
- ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007;447:799-816. [PubMed]
- Bae JB. Perspectives of international human epigenome consortium. Genomics Inform 2013;11:7-14. [PubMed]
- Bernstein BE, Stamatoyannopoulos JA, Costello JFThe NIH Roadmap Epigenomics Mapping Consortium, et al. Nat Biotechnol 2010;28:1045-8. [PubMed]
- Roadmap Epigenomics Consortium. Integrative analysis of 111 reference human epigenomes. Nature 2015;518:317-30. [PubMed]
- Skipper M, Eccleston A, Gray N, et al. Presenting the epigenome roadmap. Nature 2015;518:313. [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014;507:315-22. [PubMed]
- Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell 2013;155:462-77. [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008;455:1061-8. [PubMed]
- Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010;17:98-110. [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015;517:576-82. [PubMed]
- Davis CF, Ricketts CJ, Wang M, et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell 2014;26:319-30. [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013;499:43-9. [PubMed]
- The Cancer Genome Atlas Research Network. Genomic and Epigenomic Landscapes of Adult De Novo Acute Myeloid Leukemia. N Engl J Med 2013;368:2059-74. [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202-9. [PubMed]
- The Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell 2014;159:676-90. [PubMed]
- The Cancer Genome Atlas Research Network. Integrated genomic characterization of endometrial carcinoma. Nature 2013;497:67-73. [PubMed]
- Hoadley KA, Yau C, Wolf DM, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 2014;158:929-44. [PubMed]
- Weisenberger DJ. Characterizing DNA methylation alterations from The Cancer Genome Atlas. J Clin Invest 2014;124:17-23. [PubMed]
- The Cancer Genome Atlas Research Network. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 2013;45:1113-20. [PubMed]
- Toyota M, Ahuja N, Ohe-Toyota M, et al. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A 1999;96:8681-6. [PubMed]
- Hinoue T, Weisenberger DJ, Pan F, et al. Analysis of the association between CIMP and BRAF in colorectal cancer by DNA methylation profiling. PloS One 2009;4:e8357 [PubMed]
- Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut 2009;58:90-6. [PubMed]
- Phipps AI, Limburg PJ, Baron JA, et al. Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology 2015;148:77-87.e2.
- Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 2006;38:787-93. [PubMed]
- Weisenberger DJ, Levine AJ, Long TI, et al. Association of the colorectal CpG island methylator phenotype with molecular features, risk factors, and family history. Cancer Epidemiol Biomarkers Prev 2015;24:512-9. [PubMed]
- Ogino S, Kawasaki T, Kirkner GJ, et al. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. J Mol Diagn 2006;8:582-8. [PubMed]
- Shen L, Toyota M, Kondo Y, et al. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc Natl Acad Sci U S A 2007;104:18654-9. [PubMed]
- Cohen AL, Holmen SL, Colman H. IDH1 and IDH2 mutations in gliomas. Curr Neurol Neurosci Rep 2013;13:345. [PubMed]
- Issa JP, Kantarjian HM, Kirkpatrick P. Azacitidine. Nat Rev Drug Discov 2005;4:275-6. [PubMed]
- Christman JK. 5-Azacytidine and 5-aza-2’-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene 2002;21:5483-95. [PubMed]
- Cheng JC, Matsen CB, Gonzales FA, et al. Inhibition of DNA methylation and reactivation of silenced genes by zebularine. J Natl Cancer Inst 2003;95:399-409. [PubMed]
- Cheng JC, Yoo CB, Weisenberger DJ, et al. Preferential response of cancer cells to zebularine. Cancer Cell 2004;6:151-8. [PubMed]
- Baylin SB. Resistance, epigenetics and the cancer ecosystem. Nat Med 2011;17:288-9. [PubMed]
- Wrangle J, Wang W, Koch A, et al. Alterations of immune response of Non-Small Cell Lung Cancer with Azacytidine. Oncotarget 2013;4:2067-79. [PubMed]
- Ahuja N, Easwaran H, Baylin SB. Harnessing the potential of epigenetic therapy to target solid tumors. J Clin Invest 2014;124:56-63. [PubMed]
- Juergens RA, Wrangle J, Vendetti FP, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov 2011;1:598-607. [PubMed]
- Chuang JC, Warner SL, Vollmer D, et al. S110, a 5-Aza-2’-deoxycytidine-containing dinucleotide, is an effective DNA methylation inhibitor in vivo and can reduce tumor growth. Mol Cancer Ther 2010;9:1443-50. [PubMed]
- Yoo CB, Jeong S, Egger G, et al. Delivery of 5-aza-2’-deoxycytidine to cells using oligodeoxynucleotides. Cancer Res 2007;67:6400-8. [PubMed]


